1jch
From Proteopedia
| Line 1: | Line 1: | ||
| - | [[Image:1jch.gif|left|200px]] | + | [[Image:1jch.gif|left|200px]] |
| - | + | ||
| - | '''Crystal Structure of Colicin E3 in Complex with its Immunity Protein''' | + | {{Structure |
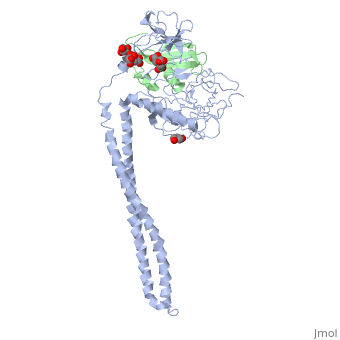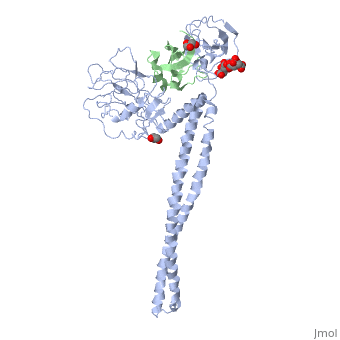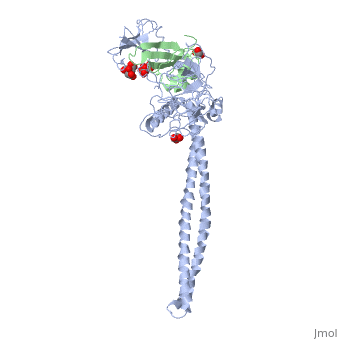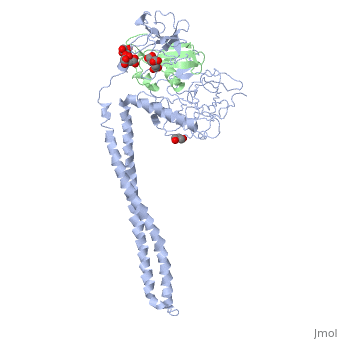
| + | |PDB= 1jch |SIZE=350|CAPTION= <scene name='initialview01'>1jch</scene>, resolution 3.02Å | ||
| + | |SITE= | ||
| + | |LIGAND= <scene name='pdbligand=CIT:CITRIC+ACID'>CIT</scene> and <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene> | ||
| + | |ACTIVITY= | ||
| + | |GENE= | ||
| + | }} | ||
| + | |||
| + | '''Crystal Structure of Colicin E3 in Complex with its Immunity Protein''' | ||
| + | |||
==Overview== | ==Overview== | ||
| Line 7: | Line 16: | ||
==About this Structure== | ==About this Structure== | ||
| - | 1JCH is a [ | + | 1JCH is a [[Protein complex]] structure of sequences from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1JCH OCA]. |
==Reference== | ==Reference== | ||
| - | Crystal structure of colicin E3: implications for cell entry and ribosome inactivation., Soelaiman S, Jakes K, Wu N, Li C, Shoham M, Mol Cell. 2001 Nov;8(5):1053-62. PMID:[http:// | + | Crystal structure of colicin E3: implications for cell entry and ribosome inactivation., Soelaiman S, Jakes K, Wu N, Li C, Shoham M, Mol Cell. 2001 Nov;8(5):1053-62. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/11741540 11741540] |
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
[[Category: Protein complex]] | [[Category: Protein complex]] | ||
| Line 24: | Line 33: | ||
[[Category: translocation domain is a beta-jellyroll]] | [[Category: translocation domain is a beta-jellyroll]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Mar 20 12:01:40 2008'' |
Revision as of 10:01, 20 March 2008
| |||||||
| , resolution 3.02Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | and | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Crystal Structure of Colicin E3 in Complex with its Immunity Protein
Overview
Colicins kill E. coli by a process that involves binding to a surface receptor, entering the cell, and, finally, intoxicating it. The lethal action of colicin E3 is a specific cleavage in the ribosomal decoding A site. The crystal structure of colicin E3, reported here in a binary complex with its immunity protein (IP), reveals a Y-shaped molecule with the receptor binding domain forming a 100 A long stalk and the two globular heads of the translocation domain (T) and the catalytic domain (C) comprising the two arms. Active site residues are D510, H513, E517, and R545. IP is buried between T and C. Rather than blocking the active site, IP prevents access of the active site to the ribosome.
About this Structure
1JCH is a Protein complex structure of sequences from Escherichia coli. Full crystallographic information is available from OCA.
Reference
Crystal structure of colicin E3: implications for cell entry and ribosome inactivation., Soelaiman S, Jakes K, Wu N, Li C, Shoham M, Mol Cell. 2001 Nov;8(5):1053-62. PMID:11741540
Page seeded by OCA on Thu Mar 20 12:01:40 2008
Categories: Escherichia coli | Protein complex | Jakes, K. | Li, C. | Shoham, M. | Soelaiman, S. | Wu, N. | CIT | GOL | The receptor-binding domain is a coiled coil | The rnase domain is a six-stranded antiparallel beta-sheet. the immunity protein is a four-stranded antiparallel beta sheet flanked by 3 helices on one side of the sheet | Translocation domain is a beta-jellyroll