Lotem haleva/test page
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
This is a default text for your page '''Lotem haleva/test page'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Lotem haleva/test page'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
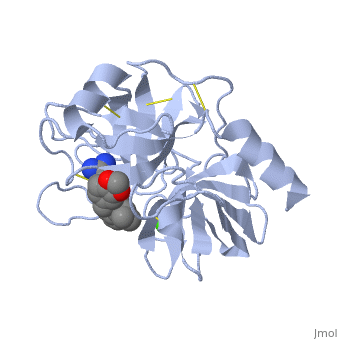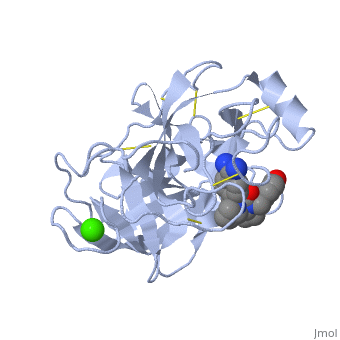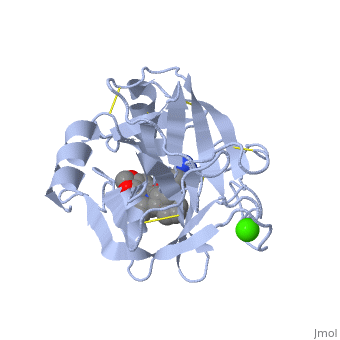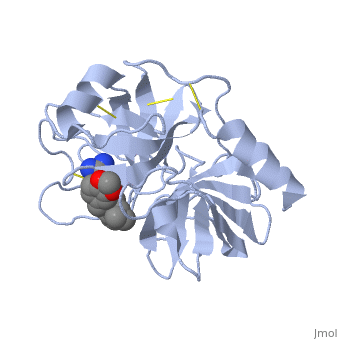
| + | is a serine protease, found in the digestive system of many vertebrates, where it hydrolyses proteins.Trypsin is produced in the pancreas as the inactive protease trypsinogen. Trypsin Break peptide chains mainly at the carboxyl side of the amino acids lysine or arginine, except when either is followed by proline. It is used for numerous biotechnological processes. The process is commonly referred to as trypsin proteolysis or trypsinisation, and proteins that have been digested/treated with trypsin are said to have been trypsinized. | ||
== Function == | == Function == | ||
| + | In the duodenum, trypsin catalyzes the hydrolysis of peptide bonds, breaking down proteins into smaller peptides. | ||
| + | The enzymatic mechanism is similar to that of other serine proteases. These enzymes contain a catalytic triad consisting of histidine-57, aspartate-102, and serine-195. These three residues form a charge relay that serves to make the active site serine nucleophilic | ||
== Disease == | == Disease == | ||
Revision as of 16:25, 22 November 2014
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644