Lotem haleva/test page
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
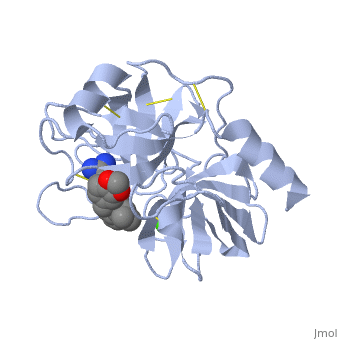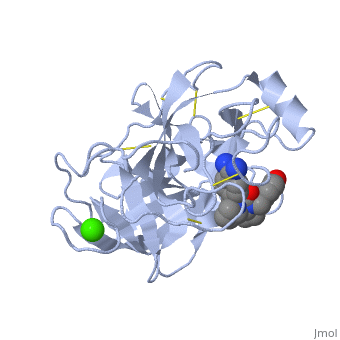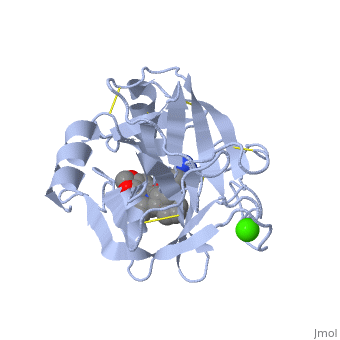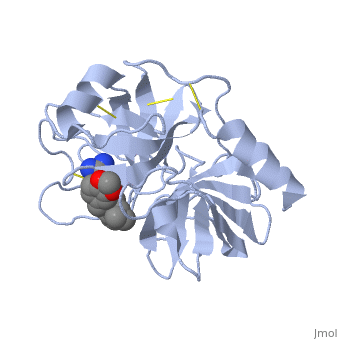
| - | is a serine protease, found in the digestive system of many vertebrates, where it hydrolyses proteins. | + | is a serine protease, found in the digestive system of many vertebrates, where it hydrolyses proteins<ref>doi:10.1016/0076-6879(94)44004-2</ref>. |
Trypsin is produced in the pancreas as the inactive protease trypsinogen. | Trypsin is produced in the pancreas as the inactive protease trypsinogen. | ||
Trypsin Break peptide chains mainly at the carboxyl side of the amino acids lysine or arginine, | Trypsin Break peptide chains mainly at the carboxyl side of the amino acids lysine or arginine, | ||
Revision as of 09:51, 23 November 2014
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Rawlings ND, Barrett AJ. Families of serine peptidases. Methods Enzymol. 1994;244:19-61. doi: 10.1016/0076-6879(94)44004-2. PMID:7845208 doi:http://dx.doi.org/10.1016/0076-6879(94)44004-2