AChE inhibitors and substrates
From Proteopedia
(Difference between revisions)
| Line 66: | Line 66: | ||
<scene name='2ack/Com_view/1'>Edrophonium (EDR)</scene> [http://en.wikipedia.org/wiki/Edrophonium] is stacked between the [http://en.wikipedia.org/wiki/Aromatic aromatic rings] of <scene name='2ack/Com_view/2'>W84 and F330</scene>, near the ''Tc''AChE <scene name='2ack/Com_view/3'>catalytic triad</scene> which consists of <font color='magenta'><b>'''S200'''</b></font>, <font color='magenta'><b>'''E327'''</b></font>, and <font color='magenta'><b>'''H440'''</b></font> ([[2ack]] or [[1ax9]]) <ref name="Ravelli">PMID:10089512</ref>. | <scene name='2ack/Com_view/1'>Edrophonium (EDR)</scene> [http://en.wikipedia.org/wiki/Edrophonium] is stacked between the [http://en.wikipedia.org/wiki/Aromatic aromatic rings] of <scene name='2ack/Com_view/2'>W84 and F330</scene>, near the ''Tc''AChE <scene name='2ack/Com_view/3'>catalytic triad</scene> which consists of <font color='magenta'><b>'''S200'''</b></font>, <font color='magenta'><b>'''E327'''</b></font>, and <font color='magenta'><b>'''H440'''</b></font> ([[2ack]] or [[1ax9]]) <ref name="Ravelli">PMID:10089512</ref>. | ||
| + | ====Galanthamine iminium derivative==== | ||
| + | The X-ray structure of ''Tc''AChE in complex with galanthamine [http://en.wikipedia.org/wiki/Iminium iminium] derivative ('''compound 5''') was determined at 2.05 Å resolution ([[1w6r]]). The <scene name='1w6r/Alignment/6'>binding mode</scene> of this compound <font color='cyan'><b>(cyan)</b></font> with ''Tc''AChE is virtually identical to that of <font color='red'><b>galanthamine (red)</b></font> itself ([[1dx6]]). The ''Tc''AChE residues which interact with galanthamine in the galanthamine/''Tc''AChE complex are colored <font color='pink'><b>pink</b></font>, while those of '''compound 5'''/''Tc''AChE are in <font color='lime'><b>lime</b></font>. The main structural change is the side-chain movement of <font color='lime'><b>Phe330</b></font> in '''compound 5'''/''Tc''AChE complex, in comparison to that of galanthamine. <font color='cyan'><b>Compound 5</b></font> differs from galanthamine by the presence of <scene name='1w6r/Alignment/7'>quaternary nitrogen atom</scene> <font color='blue'><b>(N<sup>+</sup>; blue)</b></font> instead N of <font color='red'><b>galanthamine</b></font>. This chemical difference causes the structural change in ''Tc''AChE and the slight decrease in affinity of '''compound 5''' to ''Tc''AChE in comparison to galanthamine <ref name="Greenblatt">PMID:15563167</ref>. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====Rivastigmine==== | ||
| + | Rivastigmine (Exelon) is a [http://en.wikipedia.org/wiki/Carbamate carbamate] inhibitor of AChE, and it is currenly used in therapy of [http://en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease]. [http://en.wikipedia.org/wiki/Rivastigmine Rivastigmine (Exelon)] (colored yellow) interacts with ''Tc''AChE <font color='lime'><b>(colored lime)</b></font> at the <scene name='1gqr/Active_site/4'>active-site gorge</scene> ([[1gqr]]). The carbamyl moiety of rivastigmine is <scene name='1gqr/Active_site/9'>covalently bound</scene> to the active-site S200 Oγ. The second part of rivastigmine (the leaving group), NAP ((−)-S-3-[1-(dimethylamino)ethyl]phenol) is also held in the active-site gorge, but it is <scene name='1gqr/Active_site/6'>separated</scene> from the carbamyl moiety, hence, carbamylation took place. The <scene name='1gqr/Active_site/7'>crystal structure</scene> of ''Tc''AChE/<font color='magenta'><b>NAP (colored magenta)</b></font> is known ([[1gqs]]). The <font color='violet'><b>''Tc''AChE active-site residues</b></font> which are interacting with NAP are <font color='violet'><b>colored violet</b></font>. NAP is located in a similar region of ''Tc''AChE active site, but with different orientation than that of the NAP part (colored yellow) in the ''Tc''AChE/rivastigmine complex. Only H440 and F330 significantly change their side-chain conformations. <scene name='1gqr/Active_site/8'>Overlap</scene> of the ''Tc''AChE active sites in 4 different structures (<font color='lime'><b>''Tc''AChE</b></font>/rivastigmine ([[1gqr]]), <font color='violet'><b>''Tc''AChE</b></font>/<font color='magenta'><b>NAP</b></font> ([[1gqs]]), <font color='cyan'><b>native ''Tc''AChE</b></font> ([[2ace]]), and ''Tc''AChE/'''VX''' ([[1vxr]], ''Tc''AChE colored white and VX black) reveals that the conformation of H440 in the ''Tc''AChE/NAP structure is very similar its conformation in the native ''Tc''AChE ([[2ace]]), but the distance between H440 Nδ and E327 Oε is significantly longer in the ''Tc''AChE/rivastigmine and the ''Tc''AChE/'''VX''' complexes. This structural change disrupts the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] consisting of S200, E327, H440. This could explain the very slow kinetics of AChE reactivation after its inhibition by rivastigmine <ref name="Bar-On">PMID:11888271</ref>. | ||
| + | |||
| + | {{Clear}} | ||
| + | ====Thioflavin T==== | ||
| + | The ''Tc''AChE active site consists of two binding subsites. One of them is the "catalytic anionic site" (CAS), which involves the catalytic triad <scene name='2j3q/Active_site/2'>Ser200, His440, and Glu327</scene> <font color='orange'><b>(colored orange)</b></font> and the conserved residues <scene name='2j3q/Active_site/3'>Trp84 and Phe330</scene> which also participate in ligand recognition. Another conserved residue <scene name='2j3q/Active_site/4'>Trp279</scene> <font color='cyan'><b>(colored cyan)</b></font> is situated at the second binding subsite, termed the "peripheral anionic site" (PAS), ~14 Å from CAS. <scene name='2j3q/Active_site/6'>Thioflavin T</scene> is a good example of the PAS-binding AChE inhibitors. <scene name='2j3q/Active_site/7'>Superposition</scene> of the crystal structure of the <font color='red'><b>edrophonium</b></font>/''Tc''AChE (mentioned above as a CAS-binding inhibitor) ([[2ack]]) on the <font color='magenta'><b>thioflavin T</b></font>/''Tc''AChE complex structure ([[2j3q]]) shows that these ligands' positions do not overlap. Of note is that Phe330, which is part of the CAS, is the single residue interacting with <font color='magenta'><b>thioflavin T</b></font>. This residue is the only one which significantly <scene name='2j3q/Active_site/9'>changes its conformation</scene> to avoid clashes in comparison to other CAS residues of the <font color='red'><b>edrophonium</b></font>/''Tc''AChE complex <ref name="Ravelli">PMID:10089512</ref> <ref name="Sonoda">PMID:18512913</ref>. | ||
| + | |||
| + | |||
| + | {{Clear}} | ||
| + | ====Methylene blue==== | ||
| + | The photosensitizer, <scene name='Journal:Protein_Science:1/Cv/3'>methylene blue (MB)</scene> <font color='darkmagenta'><b>(colored in darkmagenta)</b></font>, generates singlet oxygen that irreversibly inhibits Torpedo californica acetylcholinesterase (''Tc''AChE). In the dark, it inhibits reversibly. | ||
| + | MB is a noncompetitive inhibitor of ''Tc''AChE, competing with reversible inhibitors directed at both ‘‘anionic’’ subsites, but a single site is involved in inhibition. The crystal structure reveals a <scene name='Journal:Protein_Science:1/Cv1/2'>single MB stacked against Trp279 in the PAS</scene>, oriented down the gorge toward the CAS ([[2w9i]]); it is plausible that irreversible inhibition is associated with photooxidation of this residue and others within the active-site gorge. Superposition of the '''PAS regions''' of the <font color='darkmagenta'><b>MB</b></font>/''Tc''AChE ([[2w9i]]) and <font color='magenta'><b>thioflavin T</b></font>/''Tc''AChE ([[2j3q]]) complexes reveals <scene name='Journal:Protein_Science:1/Cv1/4'>similarity between positions of these ligands</scene>. As the conformation of ''Tc''AChE in the crystal structures of the two complexes is practically identical, only that of the <font color='darkmagenta'><b>MB</b></font>/''Tc''AChE structure ([[2w9i]]) is shown. The kinetic and spectroscopic data showing that inhibitors binding at the '''CAS''' can impede binding of MB are reconciled by docking studies showing that the <scene name='Journal:Protein_Science:1/Cv2/5'>conformation adopted by Phe330</scene>, midway down the gorge, in the MB/''Tc''AChE crystal structure, precludes simultaneous binding of a second MB at the CAS (<font color='blueviolet'><b>2nd MB is colored blueviolet</b></font>, <span style="color:orange;background-color:black;font-weight:bold;">Phe330 of the crystal structure is in orange</span> and <font color='indigo'><b>Phe330 of the modeled structure is in indigo</b></font>). Conversely, binding of ligands at the '''CAS''' dislodges MB from its preferred locus at the '''PAS'''. The data presented demonstrate that TcAChE is a valuable model for understanding the molecular basis of local photooxidative damage.<ref name="Paz">PMID:22674800</ref> | ||
| + | {{Clear}} | ||
| + | ====OTMA==== | ||
| + | OTMA is a nonhydrolyzable [http://en.wikipedia.org/wiki/Substrate_analog substrate analogue] of AChE. Its [http://en.wikipedia.org/wiki/Hydrolysis hydrolysis] is impossible as <scene name='2vja/Common/3'>OTMA</scene> possesses <scene name='2vja/Common/4'>carbon</scene> atom instead of the <scene name='2vja/Common/5'>ester oxygen</scene> in the AChE natural substrate ACh. Similarly to ACh, OTMA covalently binds to the ''Tc''AChE ([[2vja]]) <scene name='2vja/Active_site/1'>Ser200</scene> Oγ at the CAS. At this subsite OTMA also interacts with <scene name='2vja/Active_site/2'>Trp84, Phe330</scene> ([http://en.wikipedia.org/wiki/Cation-pi_interaction cation-π interactions]); <scene name='2vja/Active_site/3'>Glu199</scene> (electrostatic interaction); <scene name='2vja/Active_site/4'>Gly118, Gly119, and Ala201</scene> (hydrogen bonds). OTMA binds not only at CAS, but also at PAS. A second OTMA molecule interacts with <scene name='2vja/Active_site/5'>Trp279, Tyr70</scene> (cation-π interactions), and <scene name='2vja/Active_site/6'>Tyr121</scene> (weak hydrogen bond) <ref name="Colletier">PMID:18701720</ref>. Thus, this dual binding mode of OTMA with ''Tc''AChE (to CAS and PAS) could be prototypical for [[AChE bivalent inhibitors]]. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 10:07, 8 March 2015
| |||||||||||
Additional Resources
For additional information, see: Alzheimer's Disease
References
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Botti SA, Felder CE, Lifson S, Sussman JL, Silman I. A modular treatment of molecular traffic through the active site of cholinesterase. Biophys J. 1999 Nov;77(5):2430-50. PMID:10545346
- ↑ 3.0 3.1 Raves ML, Harel M, Pang YP, Silman I, Kozikowski AP, Sussman JL. Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A. Nat Struct Biol. 1997 Jan;4(1):57-63. PMID:8989325
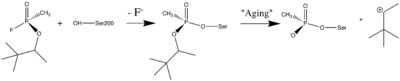
- ↑ Sanson B, Nachon F, Colletier JP, Froment MT, Toker L, Greenblatt HM, Sussman JL, Ashani Y, Masson P, Silman I, Weik M. Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime (dagger). J Med Chem. 2009 Jul 30. PMID:19642642 doi:10.1021/jm900433t
- ↑ 5.0 5.1 5.2 Millard CB, Kryger G, Ordentlich A, Greenblatt HM, Harel M, Raves ML, Segall Y, Barak D, Shafferman A, Silman I, Sussman JL. Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level. Biochemistry. 1999 Jun 1;38(22):7032-9. PMID:10353814 doi:http://dx.doi.org/10.1021/bi982678l
- ↑ Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, Axelsen PH, Silman I, Sussman JL. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9031-5. PMID:8415649
- ↑ Dvir H, Wong DM, Harel M, Barril X, Orozco M, Luque FJ, Munoz-Torrero D, Camps P, Rosenberry TL, Silman I, Sussman JL. 3D structure of Torpedo californica acetylcholinesterase complexed with huprine X at 2.1 A resolution: kinetic and molecular dynamic correlates. Biochemistry. 2002 Mar 5;41(9):2970-81. PMID:11863435
- ↑ 8.0 8.1 Greenblatt HM, Kryger G, Lewis T, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with (-)-galanthamine at 2.3 A resolution. FEBS Lett. 1999 Dec 17;463(3):321-6. PMID:10606746
- ↑ 9.0 9.1 Ravelli RB, Raves ML, Ren Z, Bourgeois D, Roth M, Kroon J, Silman I, Sussman JL. Static Laue diffraction studies on acetylcholinesterase. Acta Crystallogr D Biol Crystallogr. 1998 Nov 1;54(Pt 6 Pt 2):1359-66. PMID:10089512
- ↑ Bar-On P, Millard CB, Harel M, Dvir H, Enz A, Sussman JL, Silman I. Kinetic and structural studies on the interaction of cholinesterases with the anti-Alzheimer drug rivastigmine. Biochemistry. 2002 Mar 19;41(11):3555-64. PMID:11888271
- ↑ Harel M, Sonoda LK, Silman I, Sussman JL, Rosenberry TL. Crystal structure of thioflavin T bound to the peripheral site of Torpedo californica acetylcholinesterase reveals how thioflavin T acts as a sensitive fluorescent reporter of ligand binding to the acylation site. J Am Chem Soc. 2008 Jun 25;130(25):7856-61. Epub 2008 May 31. PMID:18512913 doi:http://dx.doi.org/10.1021/ja7109822
- ↑ Paz A, Roth E, Ashani Y, Xu Y, Shnyrov VL, Sussman JL, Silman I, Weiner L. Structural and functional characterization of the interaction of the photosensitizing probe methylene blue with Torpedo californica acetylcholinesterase. Protein Sci. 2012 Jun 1. doi: 10.1002/pro.2101. PMID:22674800 doi:10.1002/pro.2101
- ↑ Colletier JP, Bourgeois D, Sanson B, Fournier D, Sussman JL, Silman I, Weik M. Shoot-and-Trap: use of specific x-ray damage to study structural protein dynamics by temperature-controlled cryo-crystallography. Proc Natl Acad Sci U S A. 2008 Aug 19;105(33):11742-7. Epub 2008 Aug 13. PMID:18701720
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Joel L. Sussman, Michal Harel, Jaime Prilusky, David Canner