AChE inhibitors and substrates
From Proteopedia
(Difference between revisions)
| Line 108: | Line 108: | ||
====Bis(''n'')-tacrine derivatives==== | ====Bis(''n'')-tacrine derivatives==== | ||
<font color='magenta'><b>2d</b></font> and <font color='orange'><b>2f</b></font> are bis(''n'')-tacrine derivatives with n=5 and 7 (number of carbons in the linkers), respectively. These compounds are more potent and selective AChE inhibitors than tacrine alone. The binding of the tacrine moiety of <scene name='2cmf/Comparison/2'>2d</scene> at the ''Tc''AChE catalytic anionic site (CAS) is similar to that of <font color='red'><b>tacrine </b></font> in the tacrine/''Tc''AChE complex ([[1acj]]). The second tacrine moiety of the <font color='magenta'><b>2d</b></font> interacts with the peripheral anionic site (PAS) near Trp279. The interaction of <font color='magenta'><b>2d</b></font> at the CAS causes an increase of the <scene name='2cmf/Comparison/3'>distance</scene> between Ser200 Oγ and H440 Nε2 atoms, and, therefore, disruption of the catalytic triad (Ser200, H220, E327) as seen in the <font color='cyan'><b>native structure</b></font> ([[2ace]]). The binding of 2d results in <scene name='2cmf/Comparison/4'>major structural changes</scene> in the Val281-Ser291 loop changing the surface of the active-site gorge from its <font color='cyan'><b>native conformation</b></font> ([[2ace]]). The tacrine moiety of the compound <font color='orange'><b>'''2f''' (heptylene-linked bis-tacrine</b></font> at the CAS, [[2ckm]]) adopts similar <scene name='2cmf/Comparison/5'>conformation</scene> as tacrine in the tacrine/''Tc''AChE complex and the tacrine moiety of the <font color='magenta'><b>2d</b></font> at the CAS. The second tacrine moiety of the <font color='orange'><b>2f</b></font> interacts with PAS near the Trp279, like <font color='magenta'><b>2d</b></font>. The <scene name='2cmf/Comparison/6'>binding</scene> of <font color='orange'><b>2f</b></font> does not cause significant structural changes in <font color='plum'><b>''Tc''AChE</b></font> from its <font color='cyan'><b>native structure</b></font>. <scene name='2cmf/Overlap/7'>Comparison</scene> of the structures of <font color='magenta'><b>2d</b></font>/''Tc''AChE and <font color='orange'><b>2f</b></font>/''Tc''AChE reveals different contacts between the tacrine moieties of these compounds at the PAS and ''Tc''AChE. There are two additional structures of tacrine-containing ''Tc''AChE complexes: compounds <scene name='2cmf/Comparison/8'>6</scene> ([[1ut6]]) and <scene name='2cmf/Comparison/9'>7</scene> ([[1odc]]). The tacrine moieties of these compounds adopt similar conformations and interactions with CAS as the tacrine in the tacrine/''Tc''AChE, <font color='orange'><b>2f</b></font>/''Tc''AChE and <font color='magenta'><b>2d</b></font>/''Tc''AChE. Inhibitors '''6''' and '''7''' are spanning the <scene name='2cmf/Comparison/10'>active-site gorge</scene> between the CAS and the PAS, but since compound '''7''' lacks the second tacrine moiety, Trp279 adopts a different conformation in this complex structure. In the three structures: native ''Tc''AChE (cyan), '''cmp 6'''/''Tc''AChE complex (white), and '''cmp 7'''/''Tc''AChE complex (crimson) , all the ''Tc''AChE active-site gorge residues have identical conformation except Trp279 <ref name="Harel">PMID:8415649</ref> <ref name="Raves">PMID:8989325</ref> <ref name="Rydberg">PMID:16942022</ref>. | <font color='magenta'><b>2d</b></font> and <font color='orange'><b>2f</b></font> are bis(''n'')-tacrine derivatives with n=5 and 7 (number of carbons in the linkers), respectively. These compounds are more potent and selective AChE inhibitors than tacrine alone. The binding of the tacrine moiety of <scene name='2cmf/Comparison/2'>2d</scene> at the ''Tc''AChE catalytic anionic site (CAS) is similar to that of <font color='red'><b>tacrine </b></font> in the tacrine/''Tc''AChE complex ([[1acj]]). The second tacrine moiety of the <font color='magenta'><b>2d</b></font> interacts with the peripheral anionic site (PAS) near Trp279. The interaction of <font color='magenta'><b>2d</b></font> at the CAS causes an increase of the <scene name='2cmf/Comparison/3'>distance</scene> between Ser200 Oγ and H440 Nε2 atoms, and, therefore, disruption of the catalytic triad (Ser200, H220, E327) as seen in the <font color='cyan'><b>native structure</b></font> ([[2ace]]). The binding of 2d results in <scene name='2cmf/Comparison/4'>major structural changes</scene> in the Val281-Ser291 loop changing the surface of the active-site gorge from its <font color='cyan'><b>native conformation</b></font> ([[2ace]]). The tacrine moiety of the compound <font color='orange'><b>'''2f''' (heptylene-linked bis-tacrine</b></font> at the CAS, [[2ckm]]) adopts similar <scene name='2cmf/Comparison/5'>conformation</scene> as tacrine in the tacrine/''Tc''AChE complex and the tacrine moiety of the <font color='magenta'><b>2d</b></font> at the CAS. The second tacrine moiety of the <font color='orange'><b>2f</b></font> interacts with PAS near the Trp279, like <font color='magenta'><b>2d</b></font>. The <scene name='2cmf/Comparison/6'>binding</scene> of <font color='orange'><b>2f</b></font> does not cause significant structural changes in <font color='plum'><b>''Tc''AChE</b></font> from its <font color='cyan'><b>native structure</b></font>. <scene name='2cmf/Overlap/7'>Comparison</scene> of the structures of <font color='magenta'><b>2d</b></font>/''Tc''AChE and <font color='orange'><b>2f</b></font>/''Tc''AChE reveals different contacts between the tacrine moieties of these compounds at the PAS and ''Tc''AChE. There are two additional structures of tacrine-containing ''Tc''AChE complexes: compounds <scene name='2cmf/Comparison/8'>6</scene> ([[1ut6]]) and <scene name='2cmf/Comparison/9'>7</scene> ([[1odc]]). The tacrine moieties of these compounds adopt similar conformations and interactions with CAS as the tacrine in the tacrine/''Tc''AChE, <font color='orange'><b>2f</b></font>/''Tc''AChE and <font color='magenta'><b>2d</b></font>/''Tc''AChE. Inhibitors '''6''' and '''7''' are spanning the <scene name='2cmf/Comparison/10'>active-site gorge</scene> between the CAS and the PAS, but since compound '''7''' lacks the second tacrine moiety, Trp279 adopts a different conformation in this complex structure. In the three structures: native ''Tc''AChE (cyan), '''cmp 6'''/''Tc''AChE complex (white), and '''cmp 7'''/''Tc''AChE complex (crimson) , all the ''Tc''AChE active-site gorge residues have identical conformation except Trp279 <ref name="Harel">PMID:8415649</ref> <ref name="Raves">PMID:8989325</ref> <ref name="Rydberg">PMID:16942022</ref>. | ||
| + | |||
| + | ====Galantamine derivative (compound 3)==== | ||
| + | Described in the page '[[AChE inhibitors and substrates (Part II)]]' [http://en.wikipedia.org/wiki/Galantamine galanthamine] (<scene name='1w4l/Al/2'>GAL</scene>; <font color='red'><b>colored red</b></font>) is an [http://en.wikipedia.org/wiki/Acetylcholinesterase_inhibitor AChE inhibitor] and it is currently used in therapy of the [http://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's disease] (AD). Conjugate of GAL through the <scene name='1w4l/Al/3'>alkyl linker</scene> (8 carbons, <font color='black'><b>yellow</b></font>) with a <scene name='1w4l/Al/4'>phthalimido moiety</scene> <font color='blueviolet'><b>(blueviolet)</b></font> called '''compound 3''' has a larger affinity for AChE than that of GAL alone. This is similar to previously described cases of bivalent ligands. | ||
| + | A comparison between <scene name='1w4l/Comparison/1'>compound 3</scene>/''Tc''AChE ([[1w4l]]) and <scene name='1w4l/Comparison/2'>galanthamine/TcAChE</scene> structure ([[1dx6]]) shows an identical binding mode of the <font color='red'><b>GAL-moiety (transparent red)</b></font> of '''compound 3''' to that of <font color='blue'><b>GAL alone (blue)</b></font> at the CAS. A <font color='gray'><b>PEG molecule (gray)</b></font> is located at the [http://en.wikipedia.org/wiki/Active_site active site] of the galanthamine/''Tc''AChE structure. The alkyl linker spans the active-site gorge and the phthalimido moiety of '''compound 3''' is situated near Trp279 at the PAS. '''Compound 3''' has higher affinity to ''Tc''AChE than GAL. This can be explained by the higher number of interactions between '''compound 3''' (which interacts not only with residues within CAS but also within PAS) and ''Tc''AChE relative to GAL <ref name="Greenblatt">PMID:15563167</ref>. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====CPT-11==== | ||
| + | The drug <scene name='1u65/Cpt_11/1'>CPT-11</scene> <font color='black'><b>(yellow)</b></font> interacts with 13 residues of the <scene name='1u65/Binding_site/1'>active-site gorge</scene> from Trp84 at the bottom to Phe284 at the top ([[1u65]]). Nine of these residues are <scene name='1u65/Binding_site/2'>aromatic</scene> <font color='darkmagenta'><b>(Tyr70, Trp84, Tyr121, Trp279, Phe284, Phe330, Phe331, Tyr334, and His440; colored dark magenta)</b></font>. The contacts made by the drug at the bottom of the gorge involves <scene name='1u65/Binding_site/6'>complementary surface contacts</scene> with Trp84, Tyr121, Phe331, and His440 and, especially, a [http://en.wikipedia.org/wiki/Stacking_(chemistry) stacking interaction] with Phe330. The [http://en.wikipedia.org/wiki/Carbamate carbamate] moiety of [http://en.wikipedia.org/wiki/Irinotecan CPT-11] is seen near residues <scene name='1u65/Binding_site/4'>Phe331 and Tyr334</scene>. <font color='magenta'><b>Carbon C9 (shown in magenta)</b></font> of the carbamate linkage in CPT-11, is 9.3 Å from <scene name='1u65/Binding_site/5'>Ser200</scene> <font color='red'><b>Oγ, the nucleophilic atom </b></font> within the three catalytic residues Ser200, His440, and Glu327. The steric clashes between CPT-11 and ''Tc''AChE residues bar the positioning of CPT-11 near Ser200 Oγ (where [http://en.wikipedia.org/wiki/Hydrolysis hydrolysis] could occur), therefore, ''Tc''AChE can not hydrolyze CPT-11 <ref name="Harel">PMID:15772291</ref>. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====BW284C51==== | ||
| + | In a similar fashion to other AChE bivalent inhibitors, <font color='magenta'><b>BW284C51 (BW)</b></font> binds to ''Tc''AChE ([[1e3q]]) at both subsites of its <scene name='1e3q/Active_site/1'>active site</scene> - CAS and PAS. At the CAS, the BW makes a cation-aromatic interaction via its quaternary group to <scene name='1e3q/Active_site/2'>Trp84</scene> <font color='orange'><b>(colored orange)</b></font>. The BW phenyl ring forms an aromatic-aromatic interaction with His440. There is also an electrostatic interaction between the BW proximal quaternary group and Glu199. Near the PAS, BW via its distal quaternary group, interacts with <scene name='1e3q/Active_site/3'>Trp279</scene> <font color='cyan'><b>(colored cyan)</b></font> and forms an aromatic interaction with Tyr334. BW forms hydrogen bond with Tyr121 OH, and makes alkyl interactions with Phe331. The superposition of BW with two other AChE bivalent inhibitors <scene name='1e3q/Active_site/4'>DECA</scene> <font color='gray'><b>(decamethonium, colored gray, [[1acl]])</b></font> and <scene name='1e3q/Active_site/5'>E2020</scene> <font color='blueviolet'><b>(Aricept, colored blueviolet, [[1eve]])</b></font> at the ''Tc''AChE active site gorge reveals similar mode of binding. These 3 inhibitors form [http://en.wikipedia.org/wiki/Cation-pi_interaction cation-π] and π-π interactions with active-site gorge aromatic residues <scene name='1e3q/Active_site/6'>(Tyr70, Trp84, Trp279 and Phe330 or Tyr334)</scene> <font color='black'><b>(colored yellow)</b></font>. The superposition of <scene name='1e3q/Active_site/7'>DECA and E2020</scene> reveals their similar trajectory along the active site gorge, but <scene name='1e3q/Active_site/8'>BW</scene> has a different one. This results in <scene name='1e3q/Active_site/9'>different conformation of Phe330</scene>, which interacts with BW more strongly than with DECA and E2020. The conformations of the other important residues at the active site are similar in all these inhibitor complexes. | ||
| + | It has been shown experimentally that BW and E2020 bind to ''Tc''AChE approximately 100-fold stronger than DECA. These findings have several explanations: ''i)'' E2020 and BW are less flexible than DECA; ''ii)'' the aromatic groups of E2020 and BW form favourable π-π interactions with ''Tc''AChE aromatic residues, in contrast to DECA; and ''iii)'' <scene name='1e3q/Shape/3'>BW</scene> and <scene name='1e3q/Shape/2'>E2020</scene> have aromatic groups and, therefore, occupy more volume and better fit the active-site gorge, than <scene name='1e3q/Shape/4'>string-shaped DECA</scene>. Mutations at the mouse or chicken AChE residues, corresponding to the ''Tc''AChE <scene name='1e3q/Active_site/10'>Tyr70, Trp84, Trp279 and Tyr121</scene> <font color='red'><b>(colored red)</b></font>, cause significant increase of inhibition constant values for all these 3 inhibitors, supporting the notion that these residues are critical for inhibitor-AChE binding <ref name="Felder">PMID:12351819</ref> <ref name="Schalk">PMID:8415649</ref> <ref name="Kryger">PMID:10368299</ref>. | ||
| + | |||
| + | {{Clear}} | ||
| + | |||
| + | ====PEG-SH-350==== | ||
| + | <scene name='1jjb/Active_site/1'>PEG-SH-350</scene> is an untypical acetylcholinesterase inhibitor ([[1jjb]]). It consists of [http://en.wikipedia.org/wiki/Heptamer heptameric] [http://en.wikipedia.org/wiki/Polyethylene_glycol polyethylene glycol] (PEG) with a [http://en.wikipedia.org/wiki/Thiol thiol group] (SH) at the terminus. The thiol group binds close to the <scene name='1jjb/Active_site/4'>catalytic anionic site (CAS)</scene> and the second terminus binds to the <scene name='1jjb/Active_site/5'>peripheral anionic site (PAS)</scene>. PEG-SH-350 interacts with ''Torpedo californica'' acetylcholinesterase via a system of <scene name='1jjb/Active_site/6'>water molecules</scene> <font color='red'><b>(represented by oxygens colored red)</b></font>. Two out of the seven PEG-SH-350 [http://en.wikipedia.org/wiki/Ethylene_glycol ethylene glycol] units are in [http://en.wikipedia.org/wiki/Alkane_stereochemistry ''trans''] <scene name='1jjb/Active_site/7'>conformation</scene> <font color='blue'><b>(colored blue)</b></font>, while the others are in [http://en.wikipedia.org/wiki/Alkane_stereochemistry ''±gauche''] <scene name='1jjb/Active_site/8'>conformation</scene> <ref name="Koellner">PMID:12095250</ref>. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====Aricept==== | ||
| + | [http://en.wikipedia.org/wiki/Donepezil Aricept]. Among the most interesting drugs that have been designed to inhibit | ||
| + | [[acetylcholinesterase]] are those that have two binding sites that bind both the peripheral and catatylic sites simultaneously. Such drugs bind strongly and with high specificly. A good example is <scene name='Acetylcholinesterase/1eve_e2020/1'>the E2020/''Tc''AChE (Aricept) complex</scene> ([[1eve]]). It appears that the principal interaction between the aceylcholine and the enzyme is the relatively newly discovered cation-pi interaction between the cationic moiety of the substrate and the many aromatic residues lining the catalytic gorge. Unlike most | ||
| + | interatomic interactions in chemistry, cation-pi interactions are unusual in that their energy hardly changes as the cationic and aromatic ring centers distance vary between 4 and 7 Å, and for a wide variety of relative orientations of the aromatic rings. This gives the substrate an energetically smooth ride down the gorge with few bumps or barriers to impede passage down the gorge. Most acetylcholinesterases have a net negative charge and a large patch of negative potential around the entrance to the active site gorge. This may be useful to attract the positively charged acetycholine substrate to the site. As one travels down the gorge, this potential becomes increasingly more and more negative, reaching a peak at the active site at the base. Because of this potential, the peripherial site is thought to act like a substrate trap, that forces practically every molecule of substrate that reaches the peripheral site to travel down the gorge to the active site. This probably contributes greatly to the extremely rapid rate of degrading the substrate. This whole enzyme therefore acts like a brilliantly designed natural vacuum cleaner that clears the neurotransmitter out of the synapse extremely quickly. Yet to be solved, however, is how the products clear the active site rapidly, whether back through the gorge, or out a back door on the other side of the protein that quickly opens each catalytic cycle (Trp84 | ||
| + | is actually near the surface at the 'underside' of the protein). The X-ray structure of the E2020-''Tc''AChE complex shows that E2020 has a <scene name='1eve/E2020_close_up_with_84_279/13'>unique orientation</scene> along the active-site gorge, extending from the anionic subsite (<scene name='1eve/E2020_close_up_with_84lbld/7'>W84</scene>) of the active site, at the bottom, to the peripheral anionic site (<scene name='1eve/E2020_close_up_with_84_279lbld/5'>near W279</scene>), at the top, via aromatic stacking interactions with conserved aromatic acid residues. E2020 does not, however, interact directly with either the catalytic triad or the 'oxyanion hole' but only <scene name='1eve/E20_interactionshown/8'>indirectly via solvent molecules</scene> <ref name="Kryger"/>. | ||
| + | |||
| + | {{Clear}} | ||
| + | |||
| + | ====Decamethonium==== | ||
| + | Binding sites of [http://en.wikipedia.org/wiki/Pacific_electric_ray ''Torpedo californica''] [[acetylcholinesterase]] ([http://www.expasy.org/enzyme/3.1.1.7 EC 3.1.1.7]) with the bisquaternary [http://en.wikipedia.org/wiki/Ligand_(biochemistry) ligand] [http://en.wikipedia.org/wiki/Decamethonium decamethonium] (DME, [[1acl]]). DME is oriented along the <scene name='1acl/Active_site/1'>narrow gorge leading to the active site</scene>; one quaternary group is apposed to <font color='orange'><b>the indole moiety of</b></font> <scene name='1acl/Active_site/2'>W84</scene> (catalytic anionic site, CAS) and the other to <font color='cyan'><b>the indole moiety</b></font> [http://en.wikipedia.org/wiki/Indole] of <scene name='1acl/Active_site/3'>W279</scene>, near the top of the gorge, i.e. the "peripheral" anionic site (PAS). The only major conformational change in the structure of ''Tc''AChE is in the orientation of <scene name='1acl/Active_site/5'>F330</scene> <font color='purple'><b> (purple)</b></font> which lies parallel to the surface of the gorge, near the CAS of ''Tc''AChE which contains the <scene name='1acl/Active_site/4'>catalytic triad</scene> S200, E327 & H440<font color='magenta'><b> (magenta) </b></font>.<ref name="Schalk">PMID:8415649</ref> | ||
</StructureSection> | </StructureSection> | ||
Revision as of 10:15, 8 March 2015
| |||||||||||
Additional Resources
For additional information, see: Alzheimer's Disease
References
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Botti SA, Felder CE, Lifson S, Sussman JL, Silman I. A modular treatment of molecular traffic through the active site of cholinesterase. Biophys J. 1999 Nov;77(5):2430-50. PMID:10545346
- ↑ 3.0 3.1 3.2 Raves ML, Harel M, Pang YP, Silman I, Kozikowski AP, Sussman JL. Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A. Nat Struct Biol. 1997 Jan;4(1):57-63. PMID:8989325
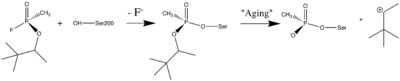
- ↑ Sanson B, Nachon F, Colletier JP, Froment MT, Toker L, Greenblatt HM, Sussman JL, Ashani Y, Masson P, Silman I, Weik M. Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime (dagger). J Med Chem. 2009 Jul 30. PMID:19642642 doi:10.1021/jm900433t
- ↑ 5.0 5.1 5.2 Millard CB, Kryger G, Ordentlich A, Greenblatt HM, Harel M, Raves ML, Segall Y, Barak D, Shafferman A, Silman I, Sussman JL. Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level. Biochemistry. 1999 Jun 1;38(22):7032-9. PMID:10353814 doi:http://dx.doi.org/10.1021/bi982678l
- ↑ 6.0 6.1 6.2 Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, Axelsen PH, Silman I, Sussman JL. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9031-5. PMID:8415649
- ↑ Dvir H, Wong DM, Harel M, Barril X, Orozco M, Luque FJ, Munoz-Torrero D, Camps P, Rosenberry TL, Silman I, Sussman JL. 3D structure of Torpedo californica acetylcholinesterase complexed with huprine X at 2.1 A resolution: kinetic and molecular dynamic correlates. Biochemistry. 2002 Mar 5;41(9):2970-81. PMID:11863435
- ↑ 8.0 8.1 8.2 Greenblatt HM, Kryger G, Lewis T, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with (-)-galanthamine at 2.3 A resolution. FEBS Lett. 1999 Dec 17;463(3):321-6. PMID:10606746
- ↑ 9.0 9.1 Ravelli RB, Raves ML, Ren Z, Bourgeois D, Roth M, Kroon J, Silman I, Sussman JL. Static Laue diffraction studies on acetylcholinesterase. Acta Crystallogr D Biol Crystallogr. 1998 Nov 1;54(Pt 6 Pt 2):1359-66. PMID:10089512
- ↑ Bar-On P, Millard CB, Harel M, Dvir H, Enz A, Sussman JL, Silman I. Kinetic and structural studies on the interaction of cholinesterases with the anti-Alzheimer drug rivastigmine. Biochemistry. 2002 Mar 19;41(11):3555-64. PMID:11888271
- ↑ Harel M, Sonoda LK, Silman I, Sussman JL, Rosenberry TL. Crystal structure of thioflavin T bound to the peripheral site of Torpedo californica acetylcholinesterase reveals how thioflavin T acts as a sensitive fluorescent reporter of ligand binding to the acylation site. J Am Chem Soc. 2008 Jun 25;130(25):7856-61. Epub 2008 May 31. PMID:18512913 doi:http://dx.doi.org/10.1021/ja7109822
- ↑ Paz A, Roth E, Ashani Y, Xu Y, Shnyrov VL, Sussman JL, Silman I, Weiner L. Structural and functional characterization of the interaction of the photosensitizing probe methylene blue with Torpedo californica acetylcholinesterase. Protein Sci. 2012 Jun 1. doi: 10.1002/pro.2101. PMID:22674800 doi:10.1002/pro.2101
- ↑ Colletier JP, Bourgeois D, Sanson B, Fournier D, Sussman JL, Silman I, Weik M. Shoot-and-Trap: use of specific x-ray damage to study structural protein dynamics by temperature-controlled cryo-crystallography. Proc Natl Acad Sci U S A. 2008 Aug 19;105(33):11742-7. Epub 2008 Aug 13. PMID:18701720
- ↑ 14.0 14.1 Haviv H, Wong DM, Greenblatt HM, Carlier PR, Pang YP, Silman I, Sussman JL. Crystal packing mediates enantioselective ligand recognition at the peripheral site of acetylcholinesterase. J Am Chem Soc. 2005 Aug 10;127(31):11029-36. PMID:16076210 doi:http://dx.doi.org/10.1021/ja051765f
- ↑ Wong DM, Greenblatt HM, Dvir H, Carlier PR, Han YF, Pang YP, Silman I, Sussman JL. Acetylcholinesterase complexed with bivalent ligands related to huperzine a: experimental evidence for species-dependent protein-ligand complementarity. J Am Chem Soc. 2003 Jan 15;125(2):363-73. PMID:12517147 doi:http://dx.doi.org/10.1021/ja021111w
- ↑ Rydberg EH, Brumshtein B, Greenblatt HM, Wong DM, Shaya D, Williams LD, Carlier PR, Pang YP, Silman I, Sussman JL. Complexes of alkylene-linked tacrine dimers with Torpedo californica acetylcholinesterase: Binding of Bis5-tacrine produces a dramatic rearrangement in the active-site gorge. J Med Chem. 2006 Sep 7;49(18):5491-500. PMID:16942022 doi:http://dx.doi.org/10.1021/jm060164b
- ↑ Felder CE, Harel M, Silman I, Sussman JL. Structure of a complex of the potent and specific inhibitor BW284C51 with Torpedo californica acetylcholinesterase. Acta Crystallogr D Biol Crystallogr. 2002 Oct;58(Pt 10 Pt 2):1765-71. Epub, 2002 Sep 28. PMID:12351819
- ↑ 18.0 18.1 Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, Axelsen PH, Silman I, Sussman JL. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9031-5. PMID:8415649
- ↑ 19.0 19.1 Kryger G, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with E2020 (Aricept): implications for the design of new anti-Alzheimer drugs. Structure. 1999 Mar 15;7(3):297-307. PMID:10368299
- ↑ Koellner G, Steiner T, Millard CB, Silman I, Sussman JL. A neutral molecule in a cation-binding site: specific binding of a PEG-SH to acetylcholinesterase from Torpedo californica. J Mol Biol. 2002 Jul 19;320(4):721-5. PMID:12095250
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Joel L. Sussman, Michal Harel, Jaime Prilusky, David Canner