Calmodulin
From Proteopedia
| Line 83: | Line 83: | ||
**[[2lqp]] - hCaM EF3 EF4 – NMR<br /> | **[[2lqp]] - hCaM EF3 EF4 – NMR<br /> | ||
**[[2kxw]] – PtapoCaM + IQ motif of NAV1.2 – NMR<br /> | **[[2kxw]] – PtapoCaM + IQ motif of NAV1.2 – NMR<br /> | ||
| - | **[[2kz2]] – CaM (mutant) – chicken<br > | + | **[[2m5e]] – PtCaM + Na channel protein<br /> |
| + | **[[2kz2]], [[4bya]] – CaM (mutant) – chicken - NMR<br > | ||
**[[2rrt]] – XlCaM (mutant) | **[[2rrt]] – XlCaM (mutant) | ||
| Line 122: | Line 123: | ||
**[[2hqw]] - rCaM+glutamate receptor peptide<br /> | **[[2hqw]] - rCaM+glutamate receptor peptide<br /> | ||
**[[3bxk]], [[3bxl]], [[4ehq]], [[4j9y]], [[4j9z]] – rCaM+ calcium channel peptide<br /> | **[[3bxk]], [[3bxl]], [[4ehq]], [[4j9y]], [[4j9z]] – rCaM+ calcium channel peptide<br /> | ||
| - | **[[1g4y]], [[3sjq]], [[4g27]], [[4g28]] - rCaM+ potassium channel CBD<br /> | + | **[[1g4y]], [[3sjq]], [[4g27]], [[4g28]], [[4qnh]] - rCaM+ potassium channel CBD<br /> |
**[[2ygg]] – rCaM + Na/H exchanger CBD<br /> | **[[2ygg]] – rCaM + Na/H exchanger CBD<br /> | ||
**[[2mgu]] – rCaM + HIV-1 matrix protein CBD – NMR<br /> | **[[2mgu]] – rCaM + HIV-1 matrix protein CBD – NMR<br /> | ||
**[[1qx7]] – rapoCaM+potassium channel peptide<br /> | **[[1qx7]] – rapoCaM+potassium channel peptide<br /> | ||
| - | **[[4gow]] – hCaM + potassium channel peptide<br /> | + | **[[4gow]], [[4umo]], [[4v0c]] – hCaM + potassium channel peptide<br /> |
**[[3hr4]], [[2ll6]], [[2ll7]] - hCaM+nitric oxide synthase CBD <br /> | **[[3hr4]], [[2ll6]], [[2ll7]] - hCaM+nitric oxide synthase CBD <br /> | ||
**[[1yr5]], [[1wrz]], [[2y4v]] – hCaM+death-associated protein kinase 1 (DAP)<br /> | **[[1yr5]], [[1wrz]], [[2y4v]] – hCaM+death-associated protein kinase 1 (DAP)<br /> | ||
| Line 132: | Line 133: | ||
**[[2m0j]], [[2m0k]] - hCaM+olfactory channel peptide<br /> | **[[2m0j]], [[2m0k]] - hCaM+olfactory channel peptide<br /> | ||
**[[2kne]] – hCaM+PMCA C-terminal CBD<br /> | **[[2kne]] – hCaM+PMCA C-terminal CBD<br /> | ||
| + | **[[4upu]] - hCaM+ IP3 3-K CBD<br /> | ||
**[[3ewt]], [[3ewv]] – hCaM+TNFR fragment<br /> | **[[3ewt]], [[3ewv]] – hCaM+TNFR fragment<br /> | ||
**[[2w73]], [[2jzi]], [[2r28]] – hCaM+Ser/Thr phosphatase CBD<br /> | **[[2w73]], [[2jzi]], [[2r28]] – hCaM+Ser/Thr phosphatase CBD<br /> | ||
| Line 151: | Line 153: | ||
**[[1ckk]] - XlCaM+CaM dependent kinase CBD - NMR<br /> | **[[1ckk]] - XlCaM+CaM dependent kinase CBD - NMR<br /> | ||
**[[2llo]], [[2llq]] – XlCaM EF-hand domain + estrogen receptor CBD – NMR<br /> | **[[2llo]], [[2llq]] – XlCaM EF-hand domain + estrogen receptor CBD – NMR<br /> | ||
| + | **[[2mes]] – XlCaM +PSD95 peptide - NMR<br /> | ||
| + | **[[2mg5]] - XlCaM+nitric oxide synthase peptide - NMR<br /> | ||
**[[2bbm]], [[2bbn]] - DmCaM+myosin light chain kinase peptide<br /> | **[[2bbm]], [[2bbn]] - DmCaM+myosin light chain kinase peptide<br /> | ||
**[[1mxe]] – DmCaM+rCaMKI CBD <br /> | **[[1mxe]] – DmCaM+rCaMKI CBD <br /> | ||
| Line 159: | Line 163: | ||
**[[4e50]] – mCam/linker/neurogranin IQ motif <br /> | **[[4e50]] – mCam/linker/neurogranin IQ motif <br /> | ||
**[[4e53]] – mCam/linker/neuromoduin IQ motif <br /> | **[[4e53]] – mCam/linker/neuromoduin IQ motif <br /> | ||
| - | **[[ | + | **[[4aqr]] – CaM-7 + calcium-transporting ATPase peptide – ''Arabidopsis thaliana''<br /> |
*Calmodulin complex with protein | *Calmodulin complex with protein | ||
| Line 169: | Line 173: | ||
**[[2lgf]] – hCaM + selectin peptide<br /> | **[[2lgf]] – hCaM + selectin peptide<br /> | ||
**[[3j41]] – hCaM + lens fiber major intrinsic protein – Cryo EM<br /> | **[[3j41]] – hCaM + lens fiber major intrinsic protein – Cryo EM<br /> | ||
| + | **[[2vas]], [[2vb6]], [[3gn4]], [[3l9i]], [[4dbp]], [[4dbq]], 2x51, 4anj – DmCaM+pMyosin IV<br /> | ||
**[[1qs7]], [[1qtx]] – CaM+RS20 – ''Escherichia coli''<br /> | **[[1qs7]], [[1qtx]] – CaM+RS20 – ''Escherichia coli''<br /> | ||
**[[1vrk]] – CaM (mutant)+RS20<br /> | **[[1vrk]] – CaM (mutant)+RS20<br /> | ||
| Line 174: | Line 179: | ||
**[[4ds7]] – CaM + spindle pole body component 10 – ''Kluyveromyces lactis''<br /> | **[[4ds7]] – CaM + spindle pole body component 10 – ''Kluyveromyces lactis''<br /> | ||
**[[2dfs]] – cCaM+myosin 5A<br /> | **[[2dfs]] – cCaM+myosin 5A<br /> | ||
| - | **[[4l79]] – hCaM + myosin 1B<br /> | + | **[[4l79]], [[4byf]] – hCaM + myosin 1B<br /> |
| + | **[[4r8g]] – rCaM + myosin 1B<br /> | ||
**[[2x0g]] – hCaM+death-associated protein kinase 1 (DAP)<br /> | **[[2x0g]] – hCaM+death-associated protein kinase 1 (DAP)<br /> | ||
| - | **[[4djc]], [[2l53]] - hCaM+ sodium channel protein type V α subunit<br /> | + | **[[4djc]], [[2l53]], [[4ovn]] - hCaM+ sodium channel protein type V α subunit<br /> |
| - | **[[4dck]] - hCaM+ sodium channel protein type V α subunit + fibroblast growth factor 13<br /> | + | **[[4dck]], [[4jpz]] - hCaM+ sodium channel protein type V α subunit + fibroblast growth factor 13<br /> |
| + | **[[4jq0]] - hCaM+ sodium channel protein type V α subunit + fibroblast growth factor 12<br /> | ||
**[[2x51]], [[4anj]] – DmCaM+pMyosin IV<br /> | **[[2x51]], [[4anj]] – DmCaM+pMyosin IV<br /> | ||
**[[2k3s]] – CaM+smoothelin-like protein 1 – NMR<br /> | **[[2k3s]] – CaM+smoothelin-like protein 1 – NMR<br /> | ||
**[[2ix7]] – apoCaM+myosin-5A<br /> | **[[2ix7]] – apoCaM+myosin-5A<br /> | ||
**[[2bki]], [[2bkh]] – CaM+myosin VI – pig<br /> | **[[2bki]], [[2bkh]] – CaM+myosin VI – pig<br /> | ||
| + | **[[4lzx]], [[4m1l]] - hCaM+ IQ domain-containing protein G<br /> | ||
| + | **[[4q57]] – hCaM N-terminal + plectin<br /> | ||
| + | **[[4q5u]] – hCaM + calcineurin<br /> | ||
}} | }} | ||
==See Also== | ==See Also== | ||
Revision as of 11:27, 9 March 2015
Calmodulin (CaM) – calcium modulated protein – regulates various protein targets. It is used by various proteins as calcium sensor and signal transducer by binding to their calcium binding domain (CBD). It undergoes conformational change upon binding Ca++ via its 4 EF hand motives and can undergo post-translational modification. More details on apo-CaM in Calcium-free Calmodulin.
Contents |
Maximum Occurrence of Calmodulin Conformations
Maximum Occurrence, a method for making rigorous numerical assessments about the maximum percent of time that a conformer of a flexible macromolecule can exist and still be compatible with the experimental data, was used to probe the conformational disorder of Calmodulin[1].
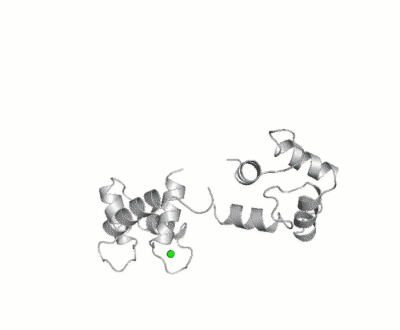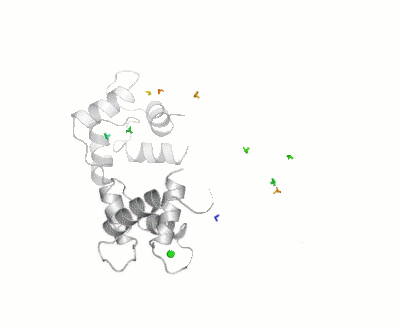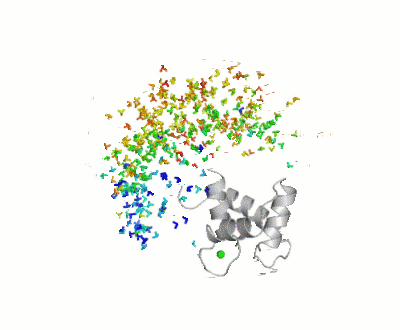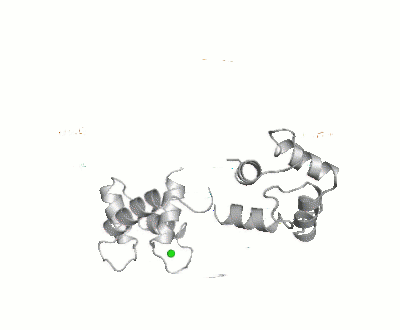
It was shown that the open (1cll) and closed (1prw) conformers can have MO of only 15% and 5% respectively.
Calmodulin in Motion
The clip represents Calmodulin in motion. At the beginning it is shown moving in the unbound form (ApoCaM), and it changes its conformation when Calcium ions are present in the medium (CaCaM).
Motion of ApoCaM is elaborated on the basis of 23 conformations derived from NMR file 1cfc, using the 3D animation program Blender, and according to a system to be published soon (Zini et al., manuscript in preparation). The transition from ApoCaM to CaCaM is elaborated with Blender starting with conformation 21 of 1cfc to arrive in conformation 11 of pdb file 1x02.
Surface rendering is also elaborated using Blender, and shows the lipophilic potential as a scale of white-black and smooth-rough, form the most lipophilic to the hydrophilic. Electrostatic potential is represented as a series of lines moving in the direction Positive to Negative, elaborated according to a scheme to be published soon (Andrei et al., in preparation). As most lines are moving towards Calmodulin, one can learn that the protein is slightly acidic (negative partial charges on its surface).
This movie was created by Andrei, Zini et al., of the Scientific Visualization Unit, Institute of Clinical Physiology - CNR of Itlay.
Conformational change of Calmodulin
,
3D Structures of Calmodulin
Updated on 09-March-2015
See Also
Bibliography
- ↑ Bertini I, Giachetti A, Luchinat C, Parigi G, Petoukhov MV, Pierattelli R, Ravera E, Svergun DI. Conformational Space of Flexible Biological Macromolecules from Average Data. J Am Chem Soc. 2010 Sep 7. PMID:20822180 doi:10.1021/ja1063923
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Karsten Theis, Jaime Prilusky, Enrico Ravera, Daniel Moyano-Marino
