Sandbox Reserved 1087
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
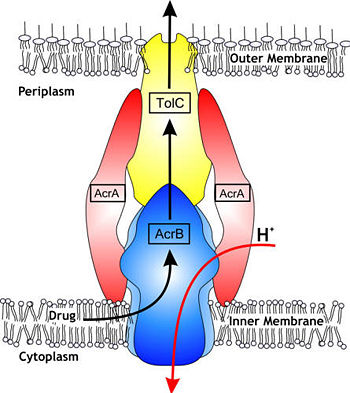
In Gram-negative bacteria, such as Escherichia coli, one of the main reasons behind bacterial multidrug resistance is resistance nodulation cell division (RND) transporters which pumps a wide range of antibiotics, dyes, bile salts and detergents out of the cell by proton motive force. Naturally, AcrB is speculated to function by pumping out of bile salts and their derivatives from the natural habitat of E. coli. for their survival with the presence of the high concentrations of these detergents. AcrB is the major RND transporter in E. coli. Three proteins form the tripartite multidrug efflux system that pumps the drugs out from the cell. The AcrB transporter, the inner membrane component of the system, cooperates with two other proteins: membrane fusion protein AcrA and an outer membrane channel TolC. The structure of the complex indicates that the drugs are transported out of the cell in a three-step procedure. [1, 2, 3] | In Gram-negative bacteria, such as Escherichia coli, one of the main reasons behind bacterial multidrug resistance is resistance nodulation cell division (RND) transporters which pumps a wide range of antibiotics, dyes, bile salts and detergents out of the cell by proton motive force. Naturally, AcrB is speculated to function by pumping out of bile salts and their derivatives from the natural habitat of E. coli. for their survival with the presence of the high concentrations of these detergents. AcrB is the major RND transporter in E. coli. Three proteins form the tripartite multidrug efflux system that pumps the drugs out from the cell. The AcrB transporter, the inner membrane component of the system, cooperates with two other proteins: membrane fusion protein AcrA and an outer membrane channel TolC. The structure of the complex indicates that the drugs are transported out of the cell in a three-step procedure. [1, 2, 3] | ||
| - | |||
| - | [[Image:a5fbdffa56.jpg]] | ||
| + | [[Image:efflux pump.jpg|thumb|left|350px|Hypothetical structure of tripartite e¥ux pumps in Gram-negative bacteria, e.g. the AcrAB^TolC e¥ux pump from E. coli]] | ||
This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| Line 41: | Line 40: | ||
== Relevance == | == Relevance == | ||
| + | |||
| + | [[Image:a5fbdffa56.jpg]] | ||
== Structural highlights == | == Structural highlights == | ||
Revision as of 10:08, 21 April 2015
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
Introduction
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644