Sandbox Reserved 1087
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
| - | == | + | == Function == |
| + | |||
| + | [[Image:Nature01050-f5.2.jpg|thumb|left|350px|text]] | ||
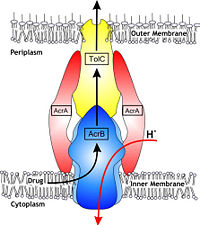
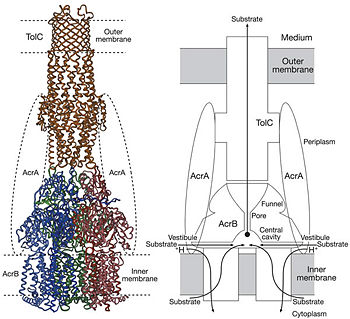
Possible mechanism of transport function has been postulated. ArcB can cooperate with TolC in the TolC docking domain forming a direct pathway from the cytoplasm to the extracellular milieu. The end of the headpiece is the narrow funnel connected with the pore, whose end is connected with the extramembrane part of the central membrane hole, namely, the cavity. There might be two pathways for the substrate translocation to the cavity. One is the groove located between TM8 and TM7 in the transmembrane domain of each promoter (Fig 1C). The other is the vestibules which are open at the side of the headpiece between PN2 and PC2 of the pore domains. The substrates located in the inner leaflet of the membrane and cytoplasm would get access to the cavity through the transmembrane groove, while the substrates which are on the outer space or in the outer leaflet of the membrane are more likely to be transported to the cavity through the vestibules. | Possible mechanism of transport function has been postulated. ArcB can cooperate with TolC in the TolC docking domain forming a direct pathway from the cytoplasm to the extracellular milieu. The end of the headpiece is the narrow funnel connected with the pore, whose end is connected with the extramembrane part of the central membrane hole, namely, the cavity. There might be two pathways for the substrate translocation to the cavity. One is the groove located between TM8 and TM7 in the transmembrane domain of each promoter (Fig 1C). The other is the vestibules which are open at the side of the headpiece between PN2 and PC2 of the pore domains. The substrates located in the inner leaflet of the membrane and cytoplasm would get access to the cavity through the transmembrane groove, while the substrates which are on the outer space or in the outer leaflet of the membrane are more likely to be transported to the cavity through the vestibules. | ||
Studies have been done on the substrate binding and specificity and the periplasmic part of the tripartite efflux system is found important to the substrate specificity. In the study on antibiotics to AcrB, Phe 386 (TM3) was reported as one the main hydrophobic contacts. However, currently, the theoretical explanation of the wide variety of substrates is still lacking. | Studies have been done on the substrate binding and specificity and the periplasmic part of the tripartite efflux system is found important to the substrate specificity. In the study on antibiotics to AcrB, Phe 386 (TM3) was reported as one the main hydrophobic contacts. However, currently, the theoretical explanation of the wide variety of substrates is still lacking. | ||
Proton-motive force has been proposed to be coupled with the tripartite efflux system. It is the transmembrane domain where binding and release of protons happen. Certain key residues had been identified as crucial to the protons translocation. They are the residues Lys940 (TM10) and Asp407 and 408 (TM4) harbored in TM 4 and 10 in each protomer. When the ion pairs between them are disrupted due to the transient protonation of residues mentioned above, there may be conformational change of TM 4 and 10. Through possible remote conformational coupling, the conformational change of TM 4 and 10 may induce the opening of the pore. | Proton-motive force has been proposed to be coupled with the tripartite efflux system. It is the transmembrane domain where binding and release of protons happen. Certain key residues had been identified as crucial to the protons translocation. They are the residues Lys940 (TM10) and Asp407 and 408 (TM4) harbored in TM 4 and 10 in each protomer. When the ion pairs between them are disrupted due to the transient protonation of residues mentioned above, there may be conformational change of TM 4 and 10. Through possible remote conformational coupling, the conformational change of TM 4 and 10 may induce the opening of the pore. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
Revision as of 10:14, 21 April 2015
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
Introduction
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644