Sandbox Reserved 1087
From Proteopedia
| Line 10: | Line 10: | ||
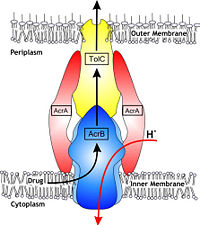
| - | In Gram-negative bacteria, such as Escherichia coli, one of the main reasons behind bacterial multidrug resistance is resistance nodulation cell division (RND) transporters which pump a wide range of dyes, bile salts, detergents and structurally unrelated antibiotics out of the cell by proton motive force driven efflux. The natural function of the efflux system is currently under debate. AcrB is the major RND transporter in E. coli. Three proteins form the tripartite multidrug efflux system that pumps the drugs out from the cell. The AcrB transporter, the inner membrane component of the system, cooperates with two other proteins: membrane fusion protein AcrA and an outer membrane channel TolC. The TolC component is connected to outer membrane and AcrB is connected to the inner membrane. The AcrA connects TolC and AcrB together (Fig 1). The structure of the complex indicates that the drugs are transported out of the cell in a three-step procedure.<ref>PMID: 12607261</ref><ref>PMID: 19166984</ref><ref>PMID: 18644451</ref> | + | In Gram-negative bacteria, such as Escherichia coli, one of the main reasons behind bacterial multidrug resistance is resistance nodulation cell division (RND) transporters which pump a wide range of dyes, bile salts, detergents and structurally unrelated antibiotics out of the cell by proton motive force driven efflux. The natural function of the efflux system is currently under debate. AcrB is the major RND transporter in E. coli. Three proteins form the tripartite multidrug efflux system that pumps the drugs out from the cell. The AcrB transporter, the inner membrane component of the system, cooperates with two other proteins: membrane fusion protein AcrA and an outer membrane channel TolC. The TolC component is connected to outer membrane and AcrB is connected to the inner membrane. The AcrA connects TolC and AcrB together (Fig 1). The structure of the complex indicates that the drugs are transported out of the cell in a three-step procedure.<ref name= "Nakashima">PMID: 12607261</ref><ref name= "Klaas">PMID: 19166984</ref><ref name= "Murasaki">PMID: 18644451</ref> |
| Line 27: | Line 27: | ||
individually coloured (blue, green and red), b) Top view of a ribbon representation, c) Structure within a slab of the transmembrane domain parallel to the membrane plane near the periplasmic surface.]] | individually coloured (blue, green and red), b) Top view of a ribbon representation, c) Structure within a slab of the transmembrane domain parallel to the membrane plane near the periplasmic surface.]] | ||
| - | In the AcrB, there is three domains: TolC docking domain, pore domain and the transmembrane domain. The AcrB consists of 1 049 amino acids and the three monomers of AcrB are organized as a trimer (Fig. 2). The first structure of the trimer was derived in symmetric manner <scene name='70/700000/Acrb_trimer/1'>trimer</scene> (Fig. 2) at 3.5 Å resolution. Later, a new 2.9 Å asymmetric conformation of trimeric AcrB was obtained and three structurally different monomers were spotted<ref>PMID: 16946072</ref>. The appearance of the trimer is of a jellyfish. The N- and C-terminal half’s show similar structural architecture and indicates at early gene duplication events. The trimer, formed by AcrB monomers, appears to be stabilized by the intermonomer connecting loops.<ref | + | In the AcrB, there is three domains: TolC docking domain, pore domain and the transmembrane domain. The AcrB consists of 1 049 amino acids and the three monomers of AcrB are organized as a trimer (Fig. 2). The first structure of the trimer was derived in symmetric manner <scene name='70/700000/Acrb_trimer/1'>trimer</scene> (Fig. 2) at 3.5 Å resolution. Later, a new 2.9 Å asymmetric conformation of trimeric AcrB was obtained and three structurally different monomers were spotted<ref>PMID: 16946072</ref>. The appearance of the trimer is of a jellyfish. The N- and C-terminal half’s show similar structural architecture and indicates at early gene duplication events. The trimer, formed by AcrB monomers, appears to be stabilized by the intermonomer connecting loops.<ref name= "Nakashima"> |
'''<scene name='70/700000/Tolc_docking/1'>TolC docking domain</scene>''' | '''<scene name='70/700000/Tolc_docking/1'>TolC docking domain</scene>''' | ||
| - | The TolC docking domain, located in the periplasm, has two subdomains: <scene name='70/700000/Acrb_dn/1'>DN</scene> and <scene name='70/700000/Acrb_dc/1'>DC</scene>. The subdomains contain a four-stranded mixed b-sheet. The TolC domain forms a funnel-like structure that has a similar diameter as the bottom of TolC. The TolC docking domain of AcrB and the bottom of TolC fits well with each other for connecting.<ref> | + | The TolC docking domain, located in the periplasm, has two subdomains: <scene name='70/700000/Acrb_dn/1'>DN</scene> and <scene name='70/700000/Acrb_dc/1'>DC</scene>. The subdomains contain a four-stranded mixed b-sheet. The TolC domain forms a funnel-like structure that has a similar diameter as the bottom of TolC. The TolC docking domain of AcrB and the bottom of TolC fits well with each other for connecting.<ref name= "Nakashima"> |
| Line 41: | Line 41: | ||
'''<scene name='70/700000/Pore_domain/1'>Pore domain </scene>''' | '''<scene name='70/700000/Pore_domain/1'>Pore domain </scene>''' | ||
| - | The pore domain consists of subdomains <scene name='70/700000/Acrb_pn1/1'>PN1</scene>, <scene name='70/700000/Acrb_pn2/1'>PN2</scene>, <scene name='70/700000/Acrb_pc1/1'>PC1</scene> and <scene name='70/700000/Acrb_pc2/1'>PC2</scene> (Fig. 3A). These subdomains have a characteristic structural motif: two b-strand–a-helix–b-strand motifs are directly repeated and sandwiched with each other. This motif forms a structure in which two a-helices are located on a four-stranded antiparallel b-sheet. Three α-helices from each PN1 subdomains form a pore in the middle of the structure (Fig. 3B). The pore connects with the bottom of the funnel-like structure of the TolC domain. The extramembrane part of the central membrane hole, namely, the central cavity is present at the proximal end of the central pore (Fig. 2c). Between PN2 and PC2, there are vestibules open at the side of the pore domain into the periplasm. They have access to the central cavity. Analysis with the AcrA has suggested that PC1 and PC2 subdomains play a role in attaching the AcrA to the complex. Studies have suggested that C-terminal domain residues <scene name='70/700000/Acrb_290-357/1'>290-357</scene> play a main role in interacting with AcrA.<ref> | + | The pore domain consists of subdomains <scene name='70/700000/Acrb_pn1/1'>PN1</scene>, <scene name='70/700000/Acrb_pn2/1'>PN2</scene>, <scene name='70/700000/Acrb_pc1/1'>PC1</scene> and <scene name='70/700000/Acrb_pc2/1'>PC2</scene> (Fig. 3A). These subdomains have a characteristic structural motif: two b-strand–a-helix–b-strand motifs are directly repeated and sandwiched with each other. This motif forms a structure in which two a-helices are located on a four-stranded antiparallel b-sheet. Three α-helices from each PN1 subdomains form a pore in the middle of the structure (Fig. 3B). The pore connects with the bottom of the funnel-like structure of the TolC domain. The extramembrane part of the central membrane hole, namely, the central cavity is present at the proximal end of the central pore (Fig. 2c). Between PN2 and PC2, there are vestibules open at the side of the pore domain into the periplasm. They have access to the central cavity. Analysis with the AcrA has suggested that PC1 and PC2 subdomains play a role in attaching the AcrA to the complex. Studies have suggested that C-terminal domain residues <scene name='70/700000/Acrb_290-357/1'>290-357</scene> play a main role in interacting with AcrA.<ref name= "Nakashima"><ref name= "Klaas"> |
| Line 47: | Line 47: | ||
'''<scene name='70/700000/Transmembrane_domain/2'>Transmembrane domain</scene>''' | '''<scene name='70/700000/Transmembrane_domain/2'>Transmembrane domain</scene>''' | ||
| - | Twelve α-helices of each monomer forms the transmembrane domain. Six α-helices in the N-terminal and the six in C-terminal are arranged symmetrically. These helices are long and they reach outside the cytoplasmic surface of the membrane. The membrane domain contains an additional extra-membrane a-helix (Ia) located between <scene name='70/700000/Acrb_tm_6/2'>TM6</scene> and <scene name='70/700000/Acrb_tm7/2'>TM7</scene> attached to the cytoplasmic membrane surface. Between <scene name='70/700000/Acrb_tm8/3'>TM8</scene> and TM7 locates a groove within the transmembrane domain of each monomer. Amino acid residues 860-868 in TM8 are in a disordered state. Through the disordered region of the top of TM8, the groove is connected with the cavity. This domain contains three functionally critical residues: Asp407, Asp408 and Lys940. The residues are shown in this <scene name='70/700000/Acrb_residues/2'>link</scene>, Asp407 as pink, Asp408 as green and Lys940 as blue. When these are mutated the whole complex loses its drug resistance.<ref | + | Twelve α-helices of each monomer forms the transmembrane domain. Six α-helices in the N-terminal and the six in C-terminal are arranged symmetrically. These helices are long and they reach outside the cytoplasmic surface of the membrane. The membrane domain contains an additional extra-membrane a-helix (Ia) located between <scene name='70/700000/Acrb_tm_6/2'>TM6</scene> and <scene name='70/700000/Acrb_tm7/2'>TM7</scene> attached to the cytoplasmic membrane surface. Between <scene name='70/700000/Acrb_tm8/3'>TM8</scene> and TM7 locates a groove within the transmembrane domain of each monomer. Amino acid residues 860-868 in TM8 are in a disordered state. Through the disordered region of the top of TM8, the groove is connected with the cavity. This domain contains three functionally critical residues: Asp407, Asp408 and Lys940. The residues are shown in this <scene name='70/700000/Acrb_residues/2'>link</scene>, Asp407 as pink, Asp408 as green and Lys940 as blue. When these are mutated the whole complex loses its drug resistance.<ref name= "Nakashima"> |
| Line 54: | Line 54: | ||
[[Image:Nature01050-f5.2.jpg|thumb|right|350px|Figure 4. Proposed model of the AcrB/AcrA/TolC complex and the schematic mechanism of multidrug export mediated by AcrAB/TolC system]] | [[Image:Nature01050-f5.2.jpg|thumb|right|350px|Figure 4. Proposed model of the AcrB/AcrA/TolC complex and the schematic mechanism of multidrug export mediated by AcrAB/TolC system]] | ||
| - | Possible mechanism of transport function has been postulated. ArcB can cooperate with TolC in the TolC docking domain forming a direct pathway from the cytoplasm to the extracellular milieu. Based on the information of structure, there might be two pathways for the substrate translocation to the cavity. One is the groove located between TM8 and TM7 in the transmembrane domain of each monoter (Fig 1C). The other is the vestibules which are between PN2 and PC2 of the pore domains. The substrates located in the inner leaflet of the membrane and cytoplasm would get access to the cavity through the transmembrane groove, while the substrates that are on the outer space or in the outer leaflet of the membrane are more likely to be transported to the cavity through the vestibules.<ref | + | Possible mechanism of transport function has been postulated. ArcB can cooperate with TolC in the TolC docking domain forming a direct pathway from the cytoplasm to the extracellular milieu. Based on the information of structure, there might be two pathways for the substrate translocation to the cavity. One is the groove located between TM8 and TM7 in the transmembrane domain of each monoter (Fig 1C). The other is the vestibules which are between PN2 and PC2 of the pore domains. The substrates located in the inner leaflet of the membrane and cytoplasm would get access to the cavity through the transmembrane groove, while the substrates that are on the outer space or in the outer leaflet of the membrane are more likely to be transported to the cavity through the vestibules.<ref name= "Nakashima"> |
| - | Studies have been done on the substrate binding and specificity and the periplasmic part of the tripartite efflux system is found important to the substrate specificity. In the study on antibiotics to AcrB, Phe<scene name='70/700000/Acrb_386/1'>386</scene> (TM3) was reported as one the main hydrophobic contacts. However, currently, the theoretical explanation of the wide variety of substrates is still lacking.<ref | + | Studies have been done on the substrate binding and specificity and the periplasmic part of the tripartite efflux system is found important to the substrate specificity. In the study on antibiotics to AcrB, Phe<scene name='70/700000/Acrb_386/1'>386</scene> (TM3) was reported as one the main hydrophobic contacts. However, currently, the theoretical explanation of the wide variety of substrates is still lacking.<ref name= "Nakashima"> |
| - | The tripartite efflux system coupled with proton-motive force across the cytoplasmic membrane. It is the binding and release of protons in the transmembrane domain that is crucial to the energy transduction.<ref | + | The tripartite efflux system coupled with proton-motive force across the cytoplasmic membrane. It is the binding and release of protons in the transmembrane domain that is crucial to the energy transduction.<ref name= "Klaas"> Certain key residues had been identified as crucial to the protons translocation. They are the residues Lys940 (TM10) and Asp407 and 408 (TM4) harbored in <scene name='70/700000/Acrb_tm_4/2'>TM4</scene> and <scene name='70/700000/Acrb_tm10/2'>TM10</scene> in each monomer. When the ion pairs between them are disrupted due to the transient protonation of residues mentioned above, there may be conformational change of TM 4 and 10. Through possible remote conformational coupling, the conformational change of TM 4 and 10 may induce the opening of the pore.<ref name= "Nakashima"> |
Revision as of 12:18, 23 April 2015
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
|
Introduction
In Gram-negative bacteria, such as Escherichia coli, one of the main reasons behind bacterial multidrug resistance is resistance nodulation cell division (RND) transporters which pump a wide range of dyes, bile salts, detergents and structurally unrelated antibiotics out of the cell by proton motive force driven efflux. The natural function of the efflux system is currently under debate. AcrB is the major RND transporter in E. coli. Three proteins form the tripartite multidrug efflux system that pumps the drugs out from the cell. The AcrB transporter, the inner membrane component of the system, cooperates with two other proteins: membrane fusion protein AcrA and an outer membrane channel TolC. The TolC component is connected to outer membrane and AcrB is connected to the inner membrane. The AcrA connects TolC and AcrB together (Fig 1). The structure of the complex indicates that the drugs are transported out of the cell in a three-step procedure.[1][2][3]
Structure
In the AcrB, there is three domains: TolC docking domain, pore domain and the transmembrane domain. The AcrB consists of 1 049 amino acids and the three monomers of AcrB are organized as a trimer (Fig. 2). The first structure of the trimer was derived in symmetric manner (Fig. 2) at 3.5 Å resolution. Later, a new 2.9 Å asymmetric conformation of trimeric AcrB was obtained and three structurally different monomers were spotted[4]. The appearance of the trimer is of a jellyfish. The N- and C-terminal half’s show similar structural architecture and indicates at early gene duplication events. The trimer, formed by AcrB monomers, appears to be stabilized by the intermonomer connecting loops.[1]