Sandbox Reserved 1087
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 31: | Line 31: | ||
'''<scene name='70/700000/Pore_domain/1'>Pore domain </scene>''' | '''<scene name='70/700000/Pore_domain/1'>Pore domain </scene>''' | ||
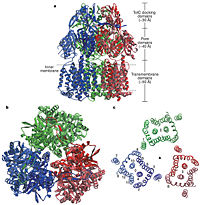
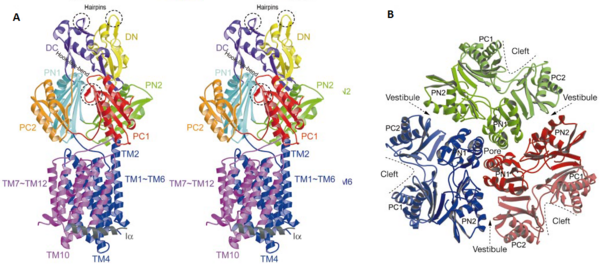
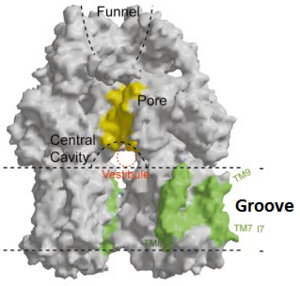
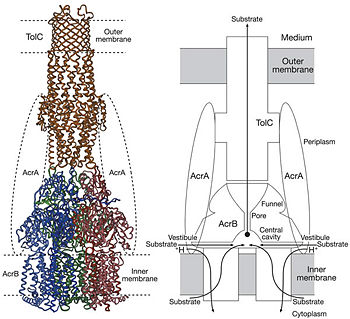
| - | The pore domain consists of subdomains <scene name='70/700000/Acrb_pn1/1'>PN1</scene>, <scene name='70/700000/Acrb_pn2/1'>PN2</scene>, <scene name='70/700000/Acrb_pc1/1'>PC1</scene> and <scene name='70/700000/Acrb_pc2/1'>PC2</scene> (Fig. 3A). These subdomains have a characteristic structural motif: two β-strand–α-helix–β-strand motifs are directly repeated and sandwiched with each other. This motif forms a structure in which two α-helices are located on a four-stranded antiparallel β-sheet. Three α-helices from each PN1 subdomains form a pore in the middle of the structure (Fig. 3B). The pore connects with the bottom of the funnel-like structure of the TolC docking domain. The extramembrane part of the central membrane hole, namely, the central cavity is present at the proximal end of the | + | The pore domain consists of subdomains <scene name='70/700000/Acrb_pn1/1'>PN1</scene>, <scene name='70/700000/Acrb_pn2/1'>PN2</scene>, <scene name='70/700000/Acrb_pc1/1'>PC1</scene> and <scene name='70/700000/Acrb_pc2/1'>PC2</scene> (Fig. 3A). These subdomains have a characteristic structural motif: two β-strand–α-helix–β-strand motifs are directly repeated and sandwiched with each other. This motif forms a structure in which two α-helices are located on a four-stranded antiparallel β-sheet. Three α-helices from each PN1 subdomains form a pore in the middle of the structure (Fig. 3B). The pore connects with the bottom of the funnel-like structure of the TolC docking domain. The extramembrane part of the central membrane hole, namely, the central cavity is present at the proximal end of the pore (Fig. 2c). Between PN2 and PC2, there are vestibules open at the side of the pore domain into the periplasm (Fig. 3B). They have access to the central cavity. Analysis with the AcrA has suggested that PC1 and PC2 subdomains are of great importance in attaching the AcrA to the complex. Studies have suggested that C-terminal domain residues <scene name='70/700000/Acrb_290-357/1'>290-357</scene> play a major role in interacting with AcrA.<ref name= "Nakashima"/><ref name= "Klaas"/> |
| Line 45: | Line 45: | ||
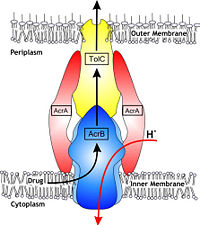
Possible mechanism of transport function has been postulated. ArcB can cooperate with TolC in the TolC docking domain forming a direct pathway from the cytoplasm to the extracellular milieu. Based on the information of structure, there might be two pathways for the substrate translocation to the central cavity. One is the groove located between TM8 and TM7 in the transmembrane domain of each monoter (Fig 1C). The other is the vestibules which are between PN2 and PC2 of the pore domains. The substrates located in the inner leaflet of the membrane and cytoplasm would get access to the central cavity through the transmembrane groove, while the substrates that are on the outer space or in the outer leaflet of the membrane are more likely to be transported to the cavity through the vestibules.<ref name= "Nakashima"/> | Possible mechanism of transport function has been postulated. ArcB can cooperate with TolC in the TolC docking domain forming a direct pathway from the cytoplasm to the extracellular milieu. Based on the information of structure, there might be two pathways for the substrate translocation to the central cavity. One is the groove located between TM8 and TM7 in the transmembrane domain of each monoter (Fig 1C). The other is the vestibules which are between PN2 and PC2 of the pore domains. The substrates located in the inner leaflet of the membrane and cytoplasm would get access to the central cavity through the transmembrane groove, while the substrates that are on the outer space or in the outer leaflet of the membrane are more likely to be transported to the cavity through the vestibules.<ref name= "Nakashima"/> | ||
| - | Studies have been done on the substrate | + | Studies have been done on the substrate specificity and the periplasmic part of the tripartite efflux system is found important to the substrate specificity. In the study on antibiotics to AcrB, Phe<scene name='70/700000/Acrb_386/1'>386</scene> (TM3) was reported as one the main hydrophobic contacts. However, currently, the theoretical explanation of the wide variety of substrates is still lacking.<ref name= "Nakashima"/> |
The tripartite efflux system coupled with proton-motive force across the cytoplasmic membrane. It is the binding and release of protons in the transmembrane domain that is crucial to the energy transduction.<ref name= "Klaas"/> Certain key residues had been identified as crucial to the protons translocation. They are the residues Lys940 (TM10) and Asp407 and 408 (TM4) harbored in <scene name='70/700000/Acrb_tm_4/2'>TM4</scene> and <scene name='70/700000/Acrb_tm10/2'>TM10</scene> in each monomer. When the ion pairs between them are disrupted due to the transient protonation of residues mentioned above, there may be conformational change of TM 4 and TM10. Through possible remote conformational coupling, the conformational change of TM 4 and TM10 may induce the opening of the pore.<ref name= "Nakashima"/> | The tripartite efflux system coupled with proton-motive force across the cytoplasmic membrane. It is the binding and release of protons in the transmembrane domain that is crucial to the energy transduction.<ref name= "Klaas"/> Certain key residues had been identified as crucial to the protons translocation. They are the residues Lys940 (TM10) and Asp407 and 408 (TM4) harbored in <scene name='70/700000/Acrb_tm_4/2'>TM4</scene> and <scene name='70/700000/Acrb_tm10/2'>TM10</scene> in each monomer. When the ion pairs between them are disrupted due to the transient protonation of residues mentioned above, there may be conformational change of TM 4 and TM10. Through possible remote conformational coupling, the conformational change of TM 4 and TM10 may induce the opening of the pore.<ref name= "Nakashima"/> | ||
Current revision
| |||||||||||