Ivan Koutsopatriy estrogen receptor
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Structural Highlights == | == Structural Highlights == | ||
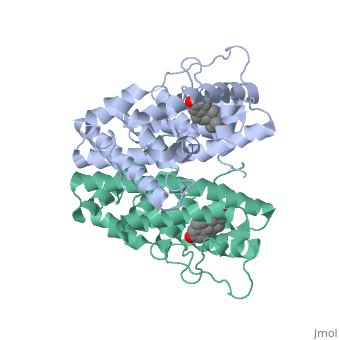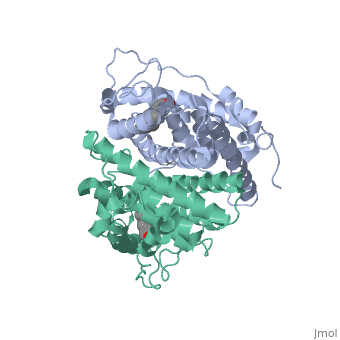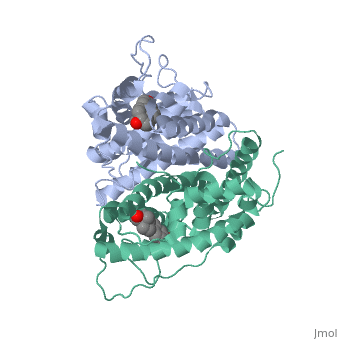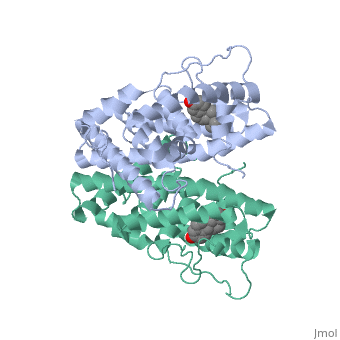
| - | ER is a modular protein composed of a ligand binding domain, a DNA binding domain and a transactivation domain. ER is a DNA-binding transcription factor. <scene name='71/714947/Er_bound_to_dna/4'>ER bound to DNA </scene> The DNA binding domain can be clearly observed in this scene; the highlighted yellow helix in close proximity to the DNA is part of the DNA binding domain. The blue beta sheet close to the yellow DNA binding alpha helix is also part of the DNA binding domain. The transactivation domain is attached at the end of the yellow DNA binding domain forming an alpha helix colored purple. The transactivation domain activates RNA polymerase when the receptor binds to DNA. The ligand binding domain may be observed here with the following scene.<scene name='71/714947/Agonist_ferutinine_bound_er/5'> Agonist ferutinine bound ER</scene> The ligand ferutinine (highlighted in pink) is bound by the ligand binding domain, composed of the surrounding blue colored alpha helices . Unbound ER normally exists loosely around the nucleus; this is subject to change depending on a multitude of factors including cell type, progress through cell cycle and reception of cellular signals. When estrogen enters the cell and binds ER, ER trans-locates and undergoes a conformation shift.<ref> Beato, M., Chavez, S., and Truss, M. (1996). Transcriptional regulation by steroid hormones. Steroids 61: 240–251. </ref> Ligand bound estrogen receptor associates more tightly with the nucleus. | + | ER is a modular protein composed of a ligand binding domain, a DNA binding domain and a transactivation domain. ER is a DNA-binding transcription factor. <scene name='71/714947/Er_bound_to_dna/4'>ER bound to DNA </scene> The DNA binding domain can be clearly observed in this scene; the highlighted yellow helix in close proximity to the DNA is part of the DNA binding domain. The blue beta sheet close to the yellow DNA binding alpha helix is also part of the DNA binding domain. The transactivation domain is attached at the end of the yellow DNA binding domain forming an alpha helix colored purple. The transactivation domain activates RNA polymerase when the receptor binds to DNA. The ligand binding domain may be observed here with the following scene.<scene name='71/714947/Agonist_ferutinine_bound_er/5'> Agonist ferutinine bound ER</scene> The ligand ferutinine (highlighted in pink) is bound by the ligand binding domain, composed of the surrounding blue colored alpha helices. Another view of the liggand binding domain is shown here, with estradiol bound. <scene name='71/714947/Er_ligand_binding_domain_estra/1'>ER ligand binding domain bound to estradiol </scene> Unbound ER normally exists loosely around the nucleus; this is subject to change depending on a multitude of factors including cell type, progress through cell cycle and reception of cellular signals. When estrogen enters the cell and binds ER, ER trans-locates and undergoes a conformation shift.<ref> Beato, M., Chavez, S., and Truss, M. (1996). Transcriptional regulation by steroid hormones. Steroids 61: 240–251. </ref> Ligand bound estrogen receptor associates more tightly with the nucleus. |
Revision as of 01:24, 31 October 2015
| |||||||||||
References
- ↑ Beato, M., Chavez, S., and Truss, M. (1996). Transcriptional regulation by steroid hormones. Steroids 61: 240–251.
- ↑ Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG (25 May 2001)."Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity.". J Biol Chem. 276 (21): 18375–83.
- ↑ Htun H, Holth LT, Walker D, Davie JR, Hager GL (1 February 1999). "Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor". Mol Biol Cell 10 (2): 471–86.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024