Ivan Koutsopatriy estrogen receptor
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
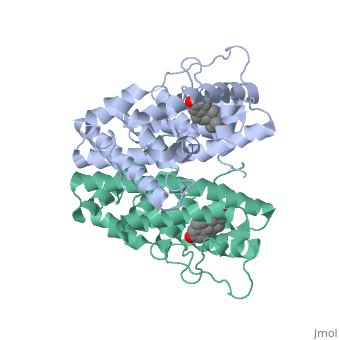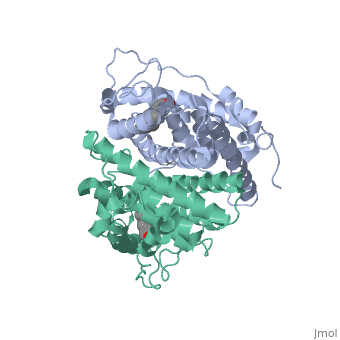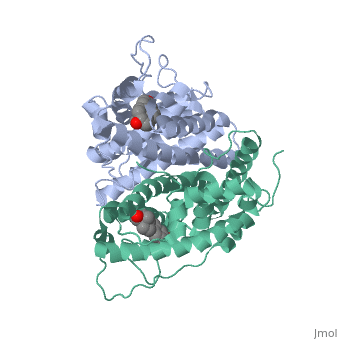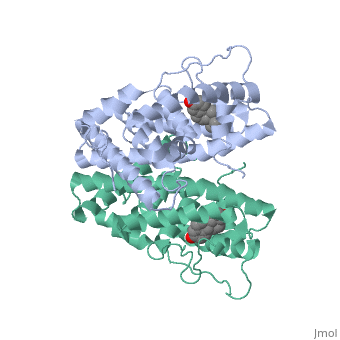
ER is functional as a ligand-dependent transcription factor. <ref> Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG (25 May 2001)."Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity.". J Biol Chem. 276 (21): 18375–83. </ref> ER responds to both agonist and antagonist ligands and can associate with the nuclear matrix. Differences in the structure of the receptor are observed depending on what ligand ER has bound (if any). Through comparisons of ER bound to agonist and antagonist ligands, some structural components may be highlighted. <scene name='71/714947/Agonist_estradiol_bound_er/2'>Agonist estradiol bound er</scene> The specific conformation of this tight loop of alpha helices and beta sheets around the ligand shows a complex capable of activating ER's transcription loci. This complex allows for the activation signal that will stimulate normal growth. | ER is functional as a ligand-dependent transcription factor. <ref> Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG (25 May 2001)."Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity.". J Biol Chem. 276 (21): 18375–83. </ref> ER responds to both agonist and antagonist ligands and can associate with the nuclear matrix. Differences in the structure of the receptor are observed depending on what ligand ER has bound (if any). Through comparisons of ER bound to agonist and antagonist ligands, some structural components may be highlighted. <scene name='71/714947/Agonist_estradiol_bound_er/2'>Agonist estradiol bound er</scene> The specific conformation of this tight loop of alpha helices and beta sheets around the ligand shows a complex capable of activating ER's transcription loci. This complex allows for the activation signal that will stimulate normal growth. | ||
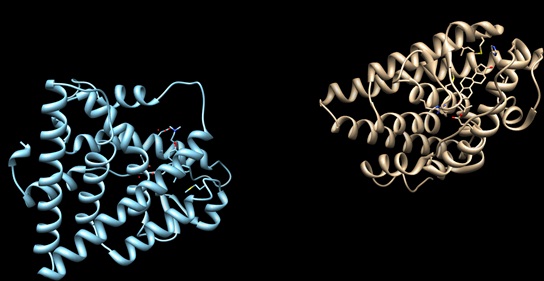
| - | Normal growth is stimulated | + | Normal growth is stimulated when an agonist bound ER binds DNA. This occurs with the assistance of chaperon proteins. These chaperons are capable of recognizing estrogen receptor ligand complexies. When ER has bound a ligand chaperons facilitate the trans-location of the complex to the nucleus. Eventually the chaperon ligand ER complex will reach specific euchromatin, at which point the chaperon protein and estrogen receptor ligand complex changes conformation so as to allow the estrogen receptor to bind the major groove at specific palindromic sequences. Estradiol is a normal ligand for ER and allows for binding in the major groove of DNA. If the ligand is an antagonist the transcription factor function of estrogen receptor becomes hindered. <scene name='71/714947/Partial_agonist_genistein_er/3'>Partial Agonist genistein bound ER</scene> The conformation of ER bound to the partial agonist genistein has a loop which is not as tight around the ligand as those found on ER with a complete agonist ligand. The ligands themselves take up different amounts of space and have varying interactions within ER. This slight difference effects the ability of the chaperon to be able to bind the receptor ligand complex to the major groove of DNA. There is a noticeable difference in the size of the pure agonist vs partial agonist scenes. Specifically, look at the width of the agonist compared to the partial agonist. Similar differences may be observed between ER which has bound the partial agonist and antagonist ligands. <scene name='71/714947/Antagonist_tamoxifen_bound_er/5'>Antagonist tamoxifen bound ER</scene> Special attention should be given to the bottom right alpha helices and beta sheets that are pushed out more in the antagonist compared to the agonist bound ER. Tamoxifen is a drug created to bind ER and inhibit the transcription factor activity of ER. Tamoxifen is larger than the normal hormone ER binds (estradiol); for this reason added with the conformation estrogen receptor takes on, the activation loop is pushed into an inactive conformation. [[Image:Left_(Blue)_antagonist_tamoxifen_bound_ER._Right_(Tan)_agonist_estradiol_bound_ER..jpg]] This blocks ER from giving the signal to grow. Antagonists are generally larger and cause estrogen receptors to be too hindered sterically to be able to bind to the major groove of DNA, inhibiting the receptor. The antagonist bound estrogen receptor is noticeably larger than the agonist bound version. Further inhibition occurs when ER has bound an antagonist ligand. Antagonist bound ER is still brought to Euchromatin in the nucleus. The larger than agonist bound ER ligand chaperon complex is not capable of binding the major groove of DNA, but still occupies the space around specific palindromic sites which ER binds and modifies the transcription of local genes to these palindromic sequence areas. This blocks agonist bound ER from being able to reach these specific palindromic major groove target loci in DNA. |
Revision as of 02:19, 11 November 2015
| |||||||||||
References
- ↑ Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG (25 May 2001)."Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity.". J Biol Chem. 276 (21): 18375–83.
- ↑ Beato, M., Chavez, S., and Truss, M. (1996). Transcriptional regulation by steroid hormones. Steroids 61: 240–251.
- ↑ Htun H, Holth LT, Walker D, Davie JR, Hager GL (1 February 1999). "Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor". Mol Biol Cell 10 (2): 471–86.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024