Glucose-fructose oxidoreductase
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
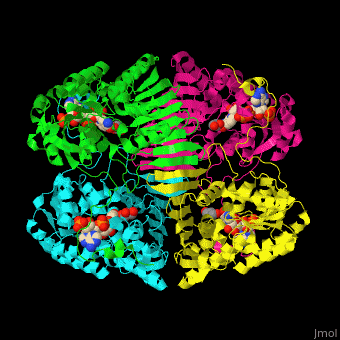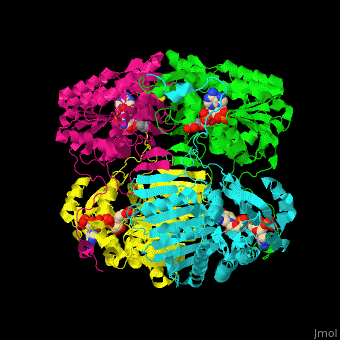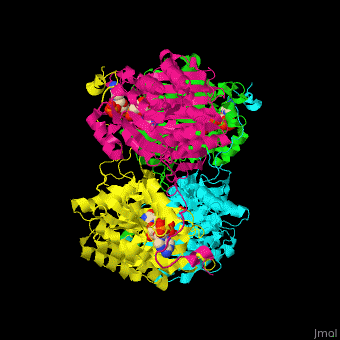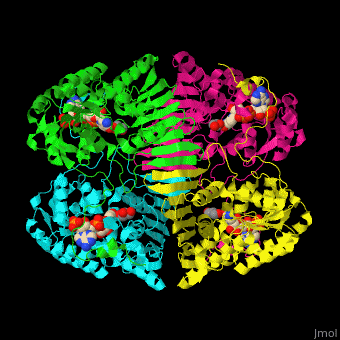
| - | The biological assembly of GFOR from ''Zymomonas mobilis'' is <scene name='49/490077/Cv/2'>homotetramer</scene> | + | The biological assembly of GFOR from ''Zymomonas mobilis'' is <scene name='49/490077/Cv/2'>homotetramer</scene>. GFOR active site contains the cofactor NADP+ and in this 3D structure the glycerol molecule seen in the active site occupies the position of the glucose ligand<ref>PMID:11705375</ref>. |
==3D structures of glucose-fructose oxidoreductase== | ==3D structures of glucose-fructose oxidoreductase== | ||
Revision as of 08:22, 13 March 2016
| |||||||||||