We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
TEM1 Class Antibiotic Resistance Proteins
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
<tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1axb|1axb]], [[1bt5|1bt5]], [[1btl|1btl]], [[1ck3|1ck3]], [[1erm|1erm]], [[1ero|1ero]], [[1erq|1erq]], [[1esu|1esu]], [[1fqg|1fqg]], [[1jtd|1jtd]], [[1jtg|1jtg]], [[1jvj|1jvj]], [[1jwp|1jwp]], [[1jwv|1jwv]], [[1jwz|1jwz]], [[1lhy|1lhy]], [[1li0|1li0]], [[1li9|1li9]], [[1m40|1m40]], [[1nxy|1nxy]], [[1ny0|1ny0]], [[1nym|1nym]], [[1nyy|1nyy]], [[1pzo|1pzo]], [[1pzp|1pzp]], [[1s0w|1s0w]], [[1tem|1tem]], [[1xxm|1xxm]], [[1yt4|1yt4]], [[1zg4|1zg4]], [[1zg6|1zg6]], [[2v1z|2v1z]]</td></tr> | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1axb|1axb]], [[1bt5|1bt5]], [[1btl|1btl]], [[1ck3|1ck3]], [[1erm|1erm]], [[1ero|1ero]], [[1erq|1erq]], [[1esu|1esu]], [[1fqg|1fqg]], [[1jtd|1jtd]], [[1jtg|1jtg]], [[1jvj|1jvj]], [[1jwp|1jwp]], [[1jwv|1jwv]], [[1jwz|1jwz]], [[1lhy|1lhy]], [[1li0|1li0]], [[1li9|1li9]], [[1m40|1m40]], [[1nxy|1nxy]], [[1ny0|1ny0]], [[1nym|1nym]], [[1nyy|1nyy]], [[1pzo|1pzo]], [[1pzp|1pzp]], [[1s0w|1s0w]], [[1tem|1tem]], [[1xxm|1xxm]], [[1yt4|1yt4]], [[1zg4|1zg4]], [[1zg6|1zg6]], [[2v1z|2v1z]]</td></tr> | ||
</table> | </table> | ||
| + | |||
| + | == Background and History == | ||
| + | Antibiotics have long been the primary line of defense against many infections. The advent of penicillin in the 1930s brought an end to an era of uncertainty: diseases once considered fatal became treatable. Unfortunately, natural selection stops for no species. Resistance to some form of antibiotic now has become standard for many infections.[A] Moreover, their liberal usage have resulted in “superbugs”, which are resistant to multiple antibiotics.[B] These strains are capable of secreting enzymes capable of deactivating the antibiotic or modifying their cell walls to render the antibiotic ineffective. | ||
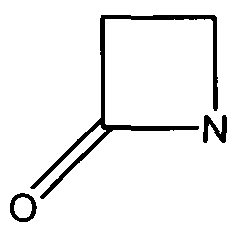
| + | Many antibiotics such as penicillin, their derivatives (penams), and cephalosporins (cephams) possess a β-lactam ring (Figure 1). | ||
| + | [[Image:Beta-Lactam-Ring.png]] | ||
| + | Figure 1. The beta-lactam ring is highlighted in red. Beta-lactamases function in hydrolyzing the amide bond within the ring, rendering the antibiotic ineffective. (Image Credit: Wikipedia) | ||
| + | |||
| + | β-Lactamases function by hydrolyzing the β-lactam ring within the antibiotic (Figure 2). This prevents the interaction between the cell wall and the antibiotic. | ||
| + | [[Image:Figure2.jpeg]] | ||
| + | Figure 2. Bacteria are capable of becoming antibiotic resistant by catalyzing the hydrolysis of the β-lactam ring. | ||
| + | |||
| + | The TEM-1 subclass is one of the most common of all β-Lactamases.[3] Their prevalence began when bacteria started exhibiting penicillin resistance on a mass scale. Since the TEM-1 subclass has been prevalent for many years, many inhibitors have either been discovered or synthesized to prevent their catalytic function. | ||
== Function == | == Function == | ||
Revision as of 22:46, 13 April 2016
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
Proteopedia Page Contributors and Editors (what is this?)
Kenna Salvatore, Matt O'Malley, Ryan Hunter Wilson, Riley Culhane, Michal Harel