CYP3A4
From Proteopedia
| Line 28: | Line 28: | ||
| - | == | + | == Relevance == |
| + | |||
| + | CYP3A4 is thought to affect the metabolism of 30-60% of the drugs on the market [https://www.pharmgkb.org/gene/PA130?tabType=tabVip#tabview=tab3&subtab=32] [http://www.pharmacytimes.com/publications/issue/2008/2008-09/2008-09-8687]. Because of this, the continual study of this enzyme is valued. A orally prescribed drug can take a while to arrive at its target cell and along the way the body has enzymes that filter out the drug dampening its efficacy. | ||
| + | |||
Revision as of 14:39, 13 May 2016
Contents |
Overview
|
Cytochrome P450 3A4 (CYP3A4) is in the heme-thiolate monooxygenase enzyme family meaning the protein contains a heme group with an iron atom. Enzymes in Cytochrome P450 are oxidizing enzymes and CYP3A4 works in the body oxidizing foreign molecules such as toxins and drugs[1][2]. It appears almost half of the marketed pharmaceutical drugs are metabolized by CYP3A4 [1] All Cytochrome P450 are essential enzymes for metabolism and the enzyme CYP3A4 is the most important. [2] Primarily found in the liver and intestine, CYP3A4 is localized in the endoplasmic reticulum membrane [3] and are present in all eukaryotic organisms as well as some prokaryotes[3]. While CYP3A4's role in drug metabolism are numerous, they are often aiding deactivation through facilitated excretion from the system or by direct inactivation. [4]
Mechanism
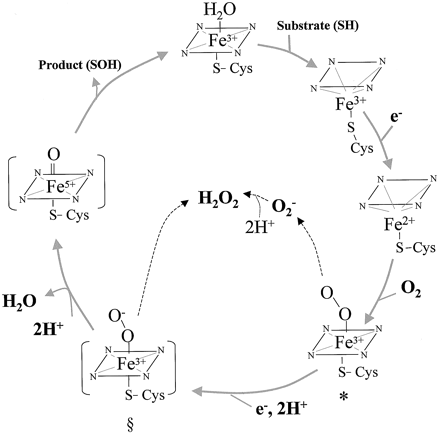
Because water molecules are stripped away when substrate binds, this causes an entropy change of 26 kcal/mol. This entropy driven reaction has a spontaneous free energy change of -7.7kcal/mol at 21ºC [4]. The cycle begins with the binding of the substrate to the ferric heme. With the addition of an electron, the heme gets reduced. This can only happen once the substrate is bound because the reduction potential is -300V which is more electronegative. After substrate binding, the induced conformational shift causes the reduction potential to be more positive at -230V making this a more thermodynamically favorable reaction. Oxygen next binds to the heme and oxidizes the iron. The addition of a second electron cleaves the oxygen-oxygen bond allowing one atom to bind two protons producing water. The second oxygen joins the substrate in what is called a monoxygenation reaction [5].
Structure and Sequence
The gene encoding CYP3A4 is located on chromosome 7 in the human genome[6] and it has been found that there are significant variants of the protein correlating to race [7]. This finding is relevant due to the proteins altered ability to react with substrates such as testosterone [8].
The
|
Relevance
CYP3A4 is thought to affect the metabolism of 30-60% of the drugs on the market [7] [8]. Because of this, the continual study of this enzyme is valued. A orally prescribed drug can take a while to arrive at its target cell and along the way the body has enzymes that filter out the drug dampening its efficacy.
External Sources
Mechanism of Oxidation Reactions Catalyzed by Cytochrome P450 Enzymes [9]
- ↑ Ogu CC, Maxa JL. Drug interactions due to cytochrome P450. Proc (Bayl Univ Med Cent). 2000 Oct;13(4):421-3. PMID:16389357
- ↑ Gonzalez FJ. Role of cytochromes P450 in chemical toxicity and oxidative stress: studies with CYP2E1. Mutat Res. 2005 Jan 6;569(1-2):101-10. PMID:15603755 doi:http://dx.doi.org/10.1016/j.mrfmmm.2004.04.021
- ↑ Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes. Chem Rev. 2004 Sep;104(9):3947-80. PMID:15352783 doi:http://dx.doi.org/10.1021/cr020443g
- ↑ Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes. Chem Rev. 2004 Sep;104(9):3947-80. PMID:15352783 doi:http://dx.doi.org/10.1021/cr020443g
- ↑ Devlin, Thomas M., ed. Textbook of Biochemistry with Clinical Correlations. 6th ed. Hoboken: John Wiley, 2006. Print.
- ↑ Inoue K, Inazawa J, Nakagawa H, Shimada T, Yamazaki H, Guengerich FP, Abe T. Assignment of the human cytochrome P-450 nifedipine oxidase gene (CYP3A4) to chromosome 7 at band q22.1 by fluorescence in situ hybridization. Jpn J Hum Genet. 1992 Jun;37(2):133-8. PMID:1391968 doi:http://dx.doi.org/10.1007/BF01899734
- ↑ Dai D, Tang J, Rose R, Hodgson E, Bienstock RJ, Mohrenweiser HW, Goldstein JA. Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J Pharmacol Exp Ther. 2001 Dec;299(3):825-31. PMID:11714865
- ↑ Dai D, Tang J, Rose R, Hodgson E, Bienstock RJ, Mohrenweiser HW, Goldstein JA. Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J Pharmacol Exp Ther. 2001 Dec;299(3):825-31. PMID:11714865