Purine repressor
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
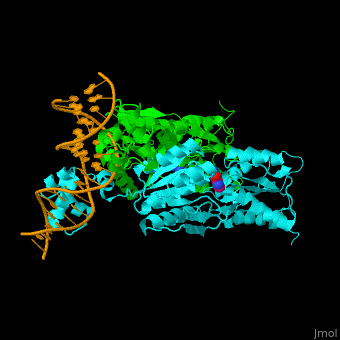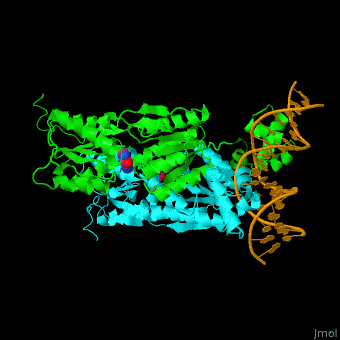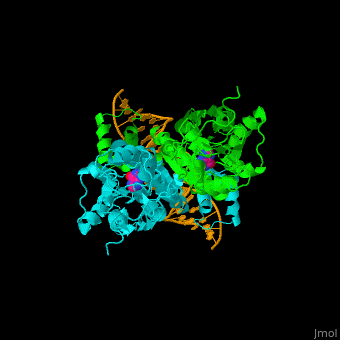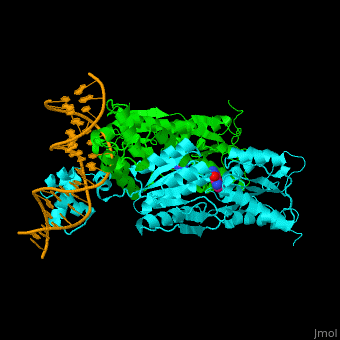
| - | <StructureSection load='1wet' size='450' side='right' caption='Structure of | + | <StructureSection load='1wet' size='450' side='right' caption='Structure of E. coli purine processor complex with DNA and guanine (PDB entry [[1wet]])' scene='55/554904/Cv/1'> |
== Function == | == Function == | ||
'''Purine repressor''' (PurR) is a member of the lac repressor family. PurP binds DNA via a highly conserved helix-turn-helix at the N terminal (DBD). PurP contains a nucleotide co-repressor binding domain as well (CBD). PurP binds to a 16-bp operator sequence and co-regulates genes which are involved in the biosynthesis of purine and pyrimidine nucleotides<ref>PMID:1971621</ref>. | '''Purine repressor''' (PurR) is a member of the lac repressor family. PurP binds DNA via a highly conserved helix-turn-helix at the N terminal (DBD). PurP contains a nucleotide co-repressor binding domain as well (CBD). PurP binds to a 16-bp operator sequence and co-regulates genes which are involved in the biosynthesis of purine and pyrimidine nucleotides<ref>PMID:1971621</ref>. | ||
Revision as of 06:36, 1 August 2016
| |||||||||||
3D structures of purine repressor
Updated on 01-August-2016
References
- ↑ Wilson HR, Turnbough CL Jr. Role of the purine repressor in the regulation of pyrimidine gene expression in Escherichia coli K-12. J Bacteriol. 1990 Jun;172(6):3208-13. PMID:1971621
- ↑ Schumacher MA, Glasfeld A, Zalkin H, Brennan RG. The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity. J Biol Chem. 1997 Sep 5;272(36):22648-53. PMID:9278422