RNA uridylyltransferase
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
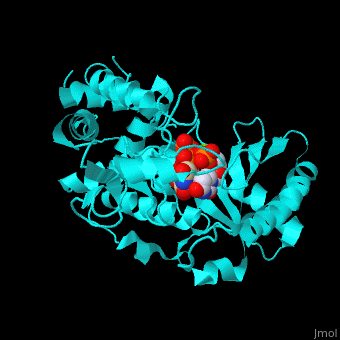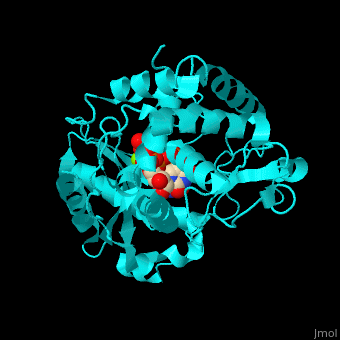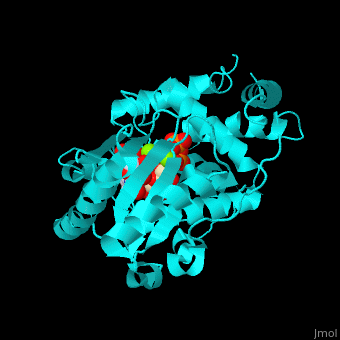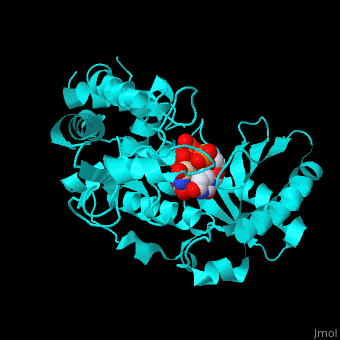
| - | RNAU-4 active site contains UTP, UMP and Mg+2 ions<ref>PMID:17785418</ref>. | + | <scene name='53/536705/Cv/3'>RNAU-4 active site contains UTP, UMP and Mg+2 ions</scene><ref>PMID:17785418</ref>. Water molecules shown as red spheres. <scene name='53/536705/Cv/4'>Mg coordination site</scene>. |
</StructureSection> | </StructureSection> | ||
Revision as of 08:46, 21 August 2016
| |||||||||||
3D Structures of RNA uridylyltransferase
Updated on 21-August-2016
References
- ↑ Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J Mol Biol. 2007 Feb 23;366(3):882-99. Epub 2006 Dec 2. PMID:17189640 doi:10.1016/j.jmb.2006.11.065
- ↑ Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc Natl Acad Sci U S A. 2007 Sep 11;104(37):14634-9. Epub 2007 Sep 4. PMID:17785418