Succinate-semialdehyde dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Structural highlights == | == Structural highlights == | ||
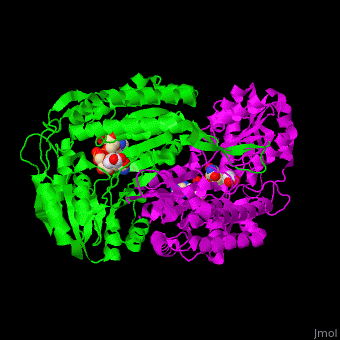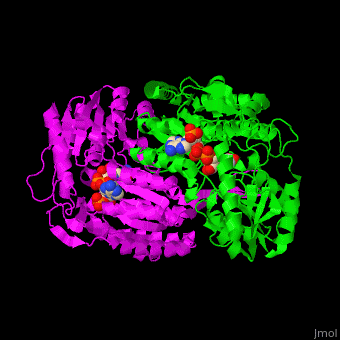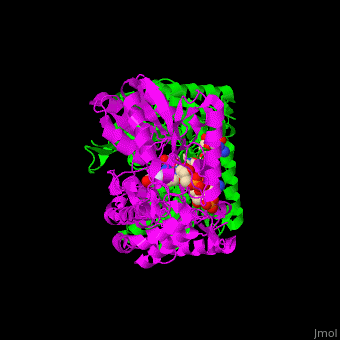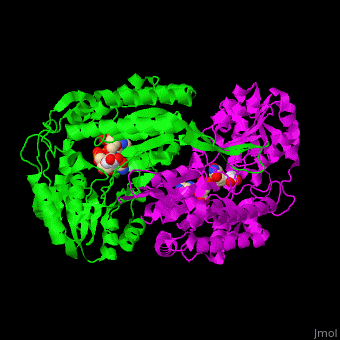
| - | The active site of SSD contains the substrate succinate semialdehyde and the cofactor NADP<ref>PMID:23500184</ref>. | + | The active site of SSD contains the <scene name='59/590825/Cv/4'>substrate succinate semialdehyde</scene> and the <scene name='59/590825/Cv/7'>cofactor NADP</scene><ref>PMID:23500184</ref>. <scene name='59/590825/Cv/9'>Whole binding site</scene>. Water molecules shown as red spheres. |
</StructureSection> | </StructureSection> | ||
Revision as of 13:17, 4 September 2016
| |||||||||||
3D structures of succinate-semialdehyde dehydrogenase
Updated on 04-September-2016
References
- ↑ JAKOBY WB, SCOTT EM. Aldehyde oxidation. III. Succinic semialdehyde dehydrogenase. J Biol Chem. 1959 Apr;234(4):937-40. PMID:13654295
- ↑ Gordon N. Succinic semialdehyde dehydrogenase deficiency (SSADH) (4-hydroxybutyric aciduria, gamma-hydroxybutyric aciduria). Eur J Paediatr Neurol. 2004;8(5):261-5. PMID:15341910 doi:http://dx.doi.org/10.1016/j.ejpn.2004.06.004
- ↑ Yuan Z, Yin B, Wei D, Yuan YR. Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase. J Struct Biol. 2013 May;182(2):125-35. doi: 10.1016/j.jsb.2013.03.001. Epub 2013 , Mar 13. PMID:23500184 doi:10.1016/j.jsb.2013.03.001