Thymidylate synthase
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Structural highlights == | == Structural highlights == | ||
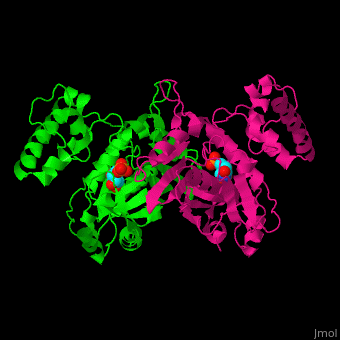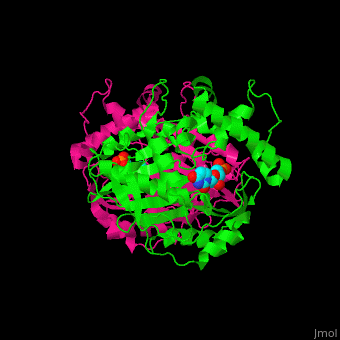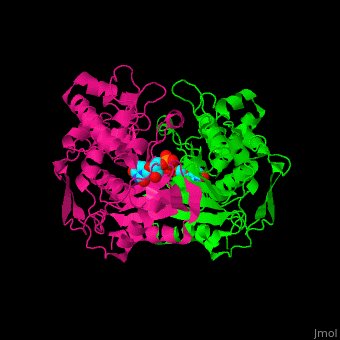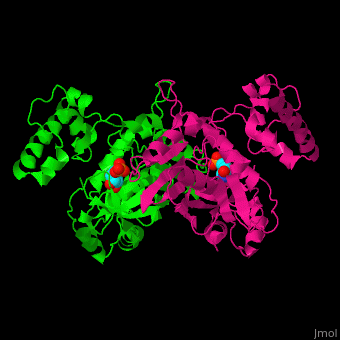
| - | TS active site contains the substrate dUMP<ref>PMID:9053905</ref>. | + | TS <scene name='49/493689/Cv/3'>active site contains the substrate dUMP</scene><ref>PMID:9053905</ref>. Water molecules shown as red spheres. |
</StructureSection> | </StructureSection> | ||
Revision as of 13:22, 14 September 2016
| |||||||||||
3D structures of thymidylate synthase
Updated on 14-September-2016
References
- ↑ Kaneda S, Nalbantoglu J, Takeishi K, Shimizu K, Gotoh O, Seno T, Ayusawa D. Structural and functional analysis of the human thymidylate synthase gene. J Biol Chem. 1990 Nov 25;265(33):20277-84. PMID:2243092
- ↑ Rahman L, Voeller D, Rahman M, Lipkowitz S, Allegra C, Barrett JC, Kaye FJ, Zajac-Kaye M. Thymidylate synthase as an oncogene: a novel role for an essential DNA synthesis enzyme. Cancer Cell. 2004 Apr;5(4):341-51. PMID:15093541
- ↑ Finer-Moore JS, Fauman EB, Morse RJ, Santi DV, Stroud RM. Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase. Protein Eng. 1996 Jan;9(1):69-75. PMID:9053905
Proteopedia Page Contributors and Editors (what is this?)
Michael O'Shaughnessy, Karsten Theis, Michal Harel, Alexander Berchansky, Shaylie Albright, Anna Postnikova, Kia Yang, Joel L. Sussman