Background
Highlights
- CRISPR-Cas9 is a powerful tool to modulate transcription in wide range of cell types.
- An expanding set of CRISPR-based transcription effectors is available.
- Gene networks can be efficiently probed and modified for biotechnology applications.[1]
CRISPR-Cas9 has recently emerged as a promising system for multiplexed genome editing as well as epigenome and transcriptome perturbation. Due to its specificity, ease of use and highly modular programmable nature, it has been widely adopted for a variety of applications such as genome editing, transcriptional inhibition and activation, genetic screening, DNA localization imaging, and many more. In this review, we will discuss non-editing applications of CRISPR-Cas9 for transcriptome perturbation, metabolic engineering, and synthetic biology.[1]
Since the early days of genetic engineering there has been a need for control of gene expression. Naturally occurring transcription factors (TFs) have traditionally been used to achieve this goal (reviewed in [2]). However, their limited DNA binding sequence space required installing specific sequences within the transcription regulatory elements of the target genes. This can be technically difficult and may have unintended consequences on gene expression. Zinc fingers (ZFs) and transcription activator-like effectors (TALEs) were developed to overcome the fixed binding sequence requirements of native TFs. However, both ZFs and TALEs have significant limitations. ZFs have complicated design criteria and large highly repetitive TALE genes are difficult to synthesize and clone (reviewed in [3][4]). These challenges have recently been overcome using CRISPR-Cas9 based TFs. The biochemical properties of CRISPR-Cas9 based TFs that enable such flexibility and describe their applications to synthetic gene circuit design and multi-plexed perturbation of native gene networks.[1]
Many bacteria and archaea possess an adaptive immune system consisting of repetitive genetic elements known as clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) proteins. Similar to RNAi pathways in eukaryotes, CRISPR–Cas systems require small RNAs for sequence-specific detection and degradation of complementary nucleic acids. Cas5 and Cas6 enzymes have evolved to specifically recognize and process CRISPR-derived transcripts into functional small RNAs used as guides by interference complexes. Our detailed understanding of these proteins has led to the development of several useful Cas6-based biotechnological methods. The structures, functions, mechanisms, and applications of the enzymes responsible for CRISPR RNA (crRNA) processing, highlighting a fascinating family of endonucleases with exquisite RNA recognition and cleavage activities are reviewed.[5]
CRISPR-Cas defense
The CRISPR-Cas systems provide protection against mobile genetic elements (MGEs) — in particular, viruses and plasmids— by sequence-specific targeting of foreign DNA or RNA [6]. A CRISPR-cas locus generally consists of an operon of CRISPR-associated (cas) genes and a CRISPR array composed of a series of direct repeats interspaced by variable DNA sequences (known as spacers) (Fig. 1A). The repeat sequences and lengths as well as the number of repeats in CRISPR arrays vary broadly, but all arrays possess the characteristic arrangement of alternating repeat and spacer sequences. The spacers are key elements of adaptive immunity, as they store the “memory” of an organism’s encounters with specific MGEs acquired as a result of a previous unsuccessful infection. This memory enables the recognition and neutralization of the invaders upon subsequent infections [6][7]. CRISPR loci are flanked by a diverse set of cas genes that define major CRISPR types based on gene conservation and locus organization[5]. Despite minimal sequence homology, Cas6s have several conserved structural features that facilitate binding of both the pre-crRNA and their crRNA product with high affinity. In most CRISPR systems, due to the pseudo-palindromic nature of the repeat sequence,the pre-crRNA adopts a stem loop structure that is bound sequence- and shape-specifically and cleaved at its base.[5] For example, PaeCas6f (Csy4) from Pseudomonas aeruginosa (2xli) in the active site. Some pre-crRNAs are predicted to be unstructured in solution and thus may be bound differently, although base pairing may be stabilized by protein interactions [8][5].
CRISPR-mediated adaptive immunity involves three steps: adaptation, expression, and interference (Fig. 1B). During the adaptation step, fragments of foreign DNA (known as protospacers) from invading elements are processed and incorporated as new spacers into the CRISPR array. The expression step involves the transcription of the CRISPR array, which is followed by processing of the precursor transcript into mature CRISPR RNAs (crRNAs)[9][7]:
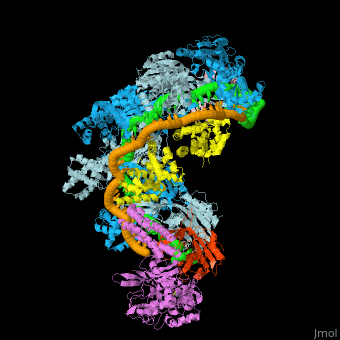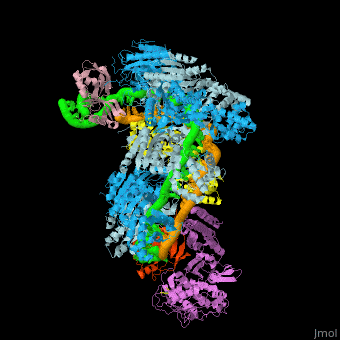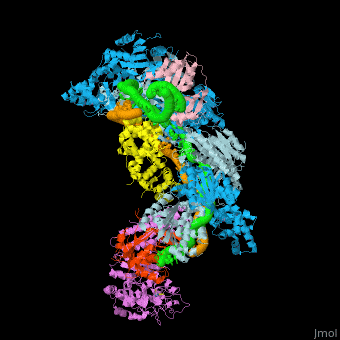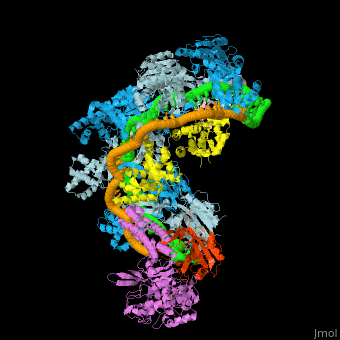
Examples of 3D structures of CRISPR RNA (crRNA)
The crRNAs are assembled with one or more Cas proteins into CRISPR ribonucleoprotein (crRNP) complexes[7].
- Example of crRNP complex with one Cas protein: (5f9r).
- Example of crRNP complex with several Cas proteins: (4qyz).
The interference step involves crRNA-directed cleavage of invading cognate virus or plasmid nucleic acids by Cas nucleases within the crRNP complex [7]. An interference complex of CRISPR-associated (Cas) proteins uses the mature crRNA as a guide to target and destroy foreign nucleic acids bearing sequence complementarity [10][9].
![Fig. 1 Overview of the CRISPR-Cas systems. (A) Architecture of class 1 (multiprotein effector complexes) and class 2 (single-protein effector complexes) CRISPR-Cas systems. (B) CRISPR-Cas adaptive immunity is mediated by CRISPR RNAs (crRNAs) and Cas proteins, which form multicomponent CRISPR ribonucleoprotein (crRNP) complexes. The first stage is adaptation, which occurs upon entry of an invading mobile genetic element (in this case, a viral genome). Cas1 (blue) and Cas2 (yellow) proteins select and process the invading DNA, and thereafter, a protospacer (orange) is integrated as a new spacer at the leader end of the CRISPR array [repeat sequences (gray) that separate similar-sized, invader-derived spacers (multiple colors)]. During the second stage, expression, the CRISPR locus is transcribed and the pre-crRNA is processed into mature crRNA guides by Cas (e.g., Cas6) or non-Cas proteins (e.g., RNase III). During the final interference stage, the Cas-crRNA complex scans invading DNA for a complementary nucleic acid target, after which the target is degraded by a Cas nuclease. From](/wiki/images/thumb/f/f2/F1r.jpg/450px-F1r.jpg)

Fig. 1 Overview of the CRISPR-Cas systems. (A) Architecture of class 1 (multiprotein effector complexes) and class 2 (single-protein effector complexes) CRISPR-Cas systems. (B) CRISPR-Cas adaptive immunity is mediated by CRISPR RNAs (crRNAs) and Cas proteins, which form multicomponent CRISPR ribonucleoprotein (crRNP) complexes. The first stage is adaptation, which occurs upon entry of an invading mobile genetic element (in this case, a viral genome). Cas1 (blue) and Cas2 (yellow) proteins select and process the invading DNA, and thereafter, a protospacer (orange) is integrated as a new spacer at the leader end of the CRISPR array [repeat sequences (gray) that separate similar-sized, invader-derived spacers (multiple colors)]. During the second stage, expression, the CRISPR locus is transcribed and the pre-crRNA is processed into mature crRNA guides by Cas (e.g., Cas6) or non-Cas proteins (e.g., RNase III). During the final interference stage, the Cas-crRNA complex scans invading DNA for a complementary nucleic acid target, after which the target is degraded by a Cas nuclease. From
[7] CRISPR-Cas diversity, classification, and evolution
The rapid evolution of highly diverse CRISPRCas systems is thought to be driven by the continuous arms race with the invading MGEs. The latest classification scheme for CRISPR-Cas systems, which takes into account the repertoire of cas genes and the sequence similarity between Cas proteins and the locus architecture, includes two classes that are currently subdivided into six types and 19 subtypes [7][11][12]. The key feature of the organization and evolution of the CRISPR-Cas loci is their pronounced modularity. The module responsible for the adaptation step is largely uniform among the diverse CRISPR-Cas systems and consists of the cas1 and cas2 genes, both of which are essential for the acquisition of spacers. In many CRISPR-Cas variants, the adaptation module also includes the cas4 gene. By contrast, the CRISPR-Cas effector module, which is involved in the maturation of the crRNAs as well as in target recognition and cleavage, shows a far greater versatility (Fig. 2A) [7][11].
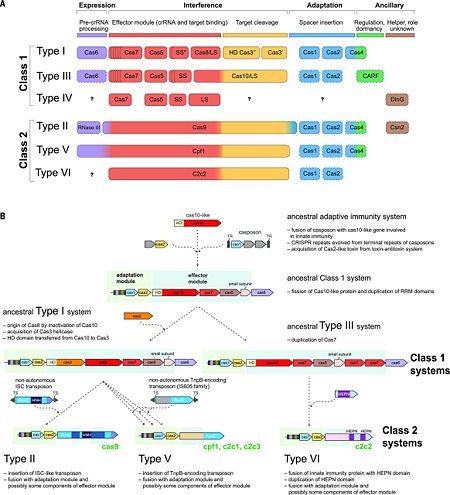
Figure. 2. CRISPR diversity and evolution. (A) Modular organization of the CRISPR-Cas systems. LS, large subunit; SS, small subunit. A putative small subunit that might be fused to the large subunit in several type I subtypes is indicated by an asterisk. Cas3 is shown as fusion of two distinct genes encoding the helicase Cas3′ and the nuclease HD Cas3′′; in some type I systems, these domains are encoded by separate genes. Functionally dispensable components are indicated by dashed outlines. Cas6 is shown with a thin solid outline for type I because it is dispensable in some systems, and by a dashed line for type III because most systems lack this gene and use the Cas6 provided in trans by other CRISPR-Cas loci. The two colors for Cas4 and C2c2 and three colors for Cas9 and Cpf1 reflect the contributions of these proteins to different stages of the CRISPR-Cas response (see text). The question marks indicate currently unknown components. From
[7][11] (B) Evolutionary scenario for the CRISPR-Cas systems. TR, terminal repeats; TS, terminal sequences; HD, HD-family endonuclease; HNH, HNH-family endonuclease; RuvC, RuvC-family endonuclease; HEPN, putative endoribonuclease of HEPN superfamily. Genes and portions of genes shown in gray denote sequences that are thought to have been encoded in the respective mobile elements but were eliminated in the course of evolution of CRISPR-Cas systems. From
[7][12]The 2 classes of CRISPR-Cas systems differ fundamentally with respect to the organization of the effector module [11]. Class 1 systems (including types I, III, and IV) are present in bacteria and archaea, and encompass effector complexes composed of 4-7 Cas protein subunits [e.g., the (CRISPR-associated complex for antiviral defense) (Cascade) of type I systems, and the Csm/Cmr complexes of type III systems]. Most of the subunits of the class 1 effector complexes — in particular, Cas5, Cas6, and Cas7—contain variants of the RNA-binding RRM (RNA recognition motif) domain. Although the sequence similarity between the individual subunits of type I and type III effector complexes is generally low, the complexes share strikingly similar overall architectures that suggest a common origin [12]. The ancestral CRISPR-Cas effector complex most likely resembled the extant type III complexes, as indicated by the presence of the archetypal type III protein, the large Cas10 subunit, which appears to be an active enzyme of the DNA polymerase–nucleotide cyclase superfamily, unlike its inactive type I counterpart (Cas8) [12]. The cas6 gene family encodes a set of RNA endonucleases responsible for crRNA processing in Type I and Type III CRISPR systems. Type II systems use a trans-activating RNA (tracrRNA) together with endogenous RNase III for crRNA maturation (Figure 2).In Type I-B, I-C, I-E, and I-F systems, the endoRNase stays bound to the crRNA and assembles into a complex with other Cas proteins for downstream targeting [9], while in Type I-A and III systems, the crRNA alone is loaded into the targeting complex and Cas6 dissociates [5].
In the less common class 2 CRISPR-Cas systems (types II, V, and VI), which are almost completely restricted to bacteria, the effector complex is represented by a single multidomain protein [11]. The best-characterized class 2 effector is Cas9 (type II), the RNA-dependent endonuclease that contains two unrelated nuclease domains, HNH and RuvC, that are responsible for the cleavage of the target and the displaced strand, respectively, in the crRNA–target DNA complex (, 4zt0). The type II loci also encode a trans-acting CRISPR RNA (tracrRNA) that evolved from the corresponding CRISPR repeat and is essential for pre-crRNA processing and target recognition in type II systems. Cas9 is directed to its DNA targets by forming a ribonucleoprotein complex with these 2 small non-coding RNAs: crRNA and tracrRNA. By elegant engineering, (4zt9[13]) that too efficiently directs Cas9 protein to DNA targets encoded within the guide sequence of sgRNA [14]:
Examples of 3D structures of single guide RNA (sgRNA)
The , termed the guide sequence, adjacent to a [14][15]. Despite this, a [14][16][17][18], more so within the 5’ proximal position of the guide sequence.
The prototype type V effector Cpf1 (subtype V-A) contains only one nuclease domain (RuvC-like) that is identifiable by sequence analysis. However, analysis of the recently solved structure of has revealed a second nuclease domain, the fold of which is unrelated to HNH or any other known nucleases. In analogy to the HNH domain in Cas9, the , and it is responsible for cleavage of the target strand.[7][19]
Screening of microbial genomes and metagenomes for undiscovered class 2 systems has resulted in the identification of three novel CRISPR-Cas variants. These include subtypes V-B and V-C, which resemble Cpf1 in that their predicted effector proteins contain a single, RuvC-like
nuclease domain. Cleavage of target DNA by the type V-B effector, denoted C2c1, has been experimentally demonstrated. Type VI is unique in that its effector protein contains two conserved HEPN domains that possess ribonuclease (RNase) activity (Fig. 2A).[7][12]
Recent comparative genomic analyses of variant CRISPR-Cas systems (Fig. 2B) [12] have revealed a strong modular evolution with multiple combinations of adaptation modules and effector modules, as well as a pivotal contribution of mobile genetic elements to the origin and diversification of the CRISPR-Cas systems. The ancestral prokaryotic adaptive immune system could have emerged via the insertion of a casposon (a recently discovered distinct class of self-synthesizing transposons that appear to encode a Cas1 homolog) next to an innate immunity locus (probably consisting of genes encoding a Cas10 nuclease and possibly one or more RNA binding proteins). Apart from providing the Cas1 nuclease/integrase that is required for recombination during spacer acquisition, the casposon may also have contributed the prototype CRISPR repeat unit that could have evolved from one of the inverted terminal repeats of the casposon. An additional toxin-antitoxin module that inserted either in the ancestral casposon or in the evolving adaptive immunity locus probably provided the cas2 gene, thus completing the adaptation module. The Cas10 nuclease and one or more additional proteins with an RRM fold (the ultimate origin of which could be a polymerase or cyclase that gave rise to Cas10) of the hybrid locus could have subsequently evolved to become the ancestral CRISPR-Cas effector module [12][7].
The widespread occurrence of class 1 systems in archaea and bacteria, together with the proliferation of the ancient RRM domain in class 1 effector proteins, strongly suggests that the ancestral CRISPR-Cas belonged to class 1. Most likely, the multiple class 2 variants then evolved via several independent replacements of the class 1 effector locus with nuclease genes that were derived from distinct MGEs (Fig. 2B). In particular, type V effector variants (Cpf1) seem to have evolved from different families of the TnpB transposase genes that are widespread in transposons [12], whereas the type II effector (Cas9) may have evolved from IscB, a protein with two nuclease domains that belongs to a recently identified distinct transposon family. Notably, class 2 CRISPR-Cas systems, in their entirety, appear to have been derived from different MGEs: Cas1 from a casposon, Cas2 from a toxin-antitoxin module, and the different effector proteins (such as Cas9 and Cpf1) from respective transposable elements [12][7].
CRISPR adaptation
The spacers of a CRISPR array represent a chronological archive of previous invader encounters. The captured spacer sequences are integrated into the CRISPR loci after exposure to MGEs, at the leader end of the array that contains the start site of CRISPR transcription. Analysis of invader target sequences (also called protospacers) has revealed a short motif directly adjacent to the target sequence, called the protospacer adjacent motif (PAM). This PAM motif allows self/nonself discrimination by the host in two ways: (i) because its presence in alien targets is required for nonself interference, and (ii) because its absence in the host’s CRISPR array avoids self-targeting. In class 1–type I and class 2–type II systems, the PAM is not only involved in interference, but also plays a role in spacer selection during the adaptation stage, implying the acquisition of functional spacers only. The PAM is a short [2 to 7 nucleotides (nt)], partially redundant sequence that in itself cannot preclude incorporation of spacers from the host DNA because of the low information content of the motif. The short PAM appears to be the result of an evolutionary trade-off between efficient incorporation of spacers from nonself DNA and preventing an autoimmune reaction.[7]
Examples of PAM:
Although host chromosomal fragments can be incorporated as new CRISPR spacers, detection of such events obviously implies that this did not result in a lethal phenotype, either due to a modified PAM and/or to an inactivated CRISPR-Cas effector module [20]. Indeed,
in the absence of the effector module, elevated
frequencies of self-spacer acquisition occur in
Escherichia coli (52). Similarly, Streptococcus
thermophilus with a catalytically inactive Cas9
results in a major increase of spacers derived
from the host genome (53). In addition, there is
a strong preference for the integration of plasmid
over chromosomal spacer sequences (52, 54, 55),
with plasmid sequences incorporated more frequently
than host DNA by two to three orders
of magnitude (56). Spacer acquisition in E. coli
requires active replication of the protospacercontaining
DNA (56). Thus, small, fast-replicating
plasmid genomes are a much better source of
spacers than the large host DNA, and such findings
are consistent with acquisition of spacers
from an infecting virus genome in the archaeon
Sulfolobus islandicus requiring its active replication
(57). In E. coli, the CRISPR-Cas system
derives the spacers primarily from products of
RecBCD-catalyzed DNA degradation that are
formed during the repair of double-stranded
breaks associated with stalled replication forks
(58). Other possible sources of substrates for
CRISPR adaptation include DNA fragments generated
either by other defense systems, such as
restriction-modification systems (59), or by the
CRISPR-Cas system itself (49).
Summary of the most extensively characterized CRISPR endoribonucleases[5][7]
Representatives of class 1 and class 2
Class 1 CRISPR-Cas systems are considered to be the evolutionary ancestral systems. The class 2 systems have evolved from class 1 systems via the insertion of transposable elements encoding various nucleases, and are now being used as tools for genome editing.[7]
(4qyz).
(3x1l).
(5f9r).
5b43.
Class 1
CRISPR subtype I-A (Cascade)
- PhoCas6nc (Cas6a) from Pyrococcus horikoshii (3qjl). . Other representatives: 3qjj, 3qjp. [21]
- SsoCas6-1A (Sso2004, Cas6-1 family, SsCas6) from Sulfolobus solfataricus (4ill). . Other representatives: 4ilm, 4ilr. [22]
- SsoCas6-1B (Sso1437, Cas6-1 family, SsoCas6) from Sulfolobus solfataricus (3zfv). . [23].
CRISPR subtype I-B (Cascade)
- MmaCas6b (Mm Cas6b) from Methanococcus maripaludis. (4z7k).
CRISPR subtype I-B?
- TthCas6B (TTHB231) from Thermus thermophilus. (4c9d). [24]. Other representative: 4c98.
CRISPR subtype I-C (Cascade)
- Cas5c (Cas5d): 4f3m (Bacillus halodurans), 3kg4 (Mannheimia succiniciproducens), 3vzh (Streptococcus pyogenes), 3vzi (Xanthomonas oryzae)[5].
CRISPR subtype I-F (Cascade)
CRISPR subtype I-U
CRISPR subtype III-A (Csm complex)
- in the type III-A CRISPR-Cas interference complex from Methanocaldococcus jannaschii.
CRISPR subtype III-B (Cmr complex)
- from Pyrococcus furiosus and Archaeoglobus fulgidus (3x1l).
- PfuCas6-1 (PfCas6) from Pyrococcus furiosus. (3pkm). Other representative: 3i4h.
CRISPR subtype Orphan
- TthCas6A (TTHB78) from T. thermophilus. (4c8z). Other representatives: 4c8y, 4c97.
CRISPR subtype IV (Csf1)
Class 2
CRISPR subtype II - SEE CRISPR-Cas9
CRISPR subtype V (Cpf1)
- from Acidaminococcus sp. BV3L6 5b43. Other representive: 5kk5.
- from Lachnospiraceae bacterium ND2006 5id6.
CRISPR subtype VI (C2c2)
See aslo
![Fig. 1 Overview of the CRISPR-Cas systems. (A) Architecture of class 1 (multiprotein effector complexes) and class 2 (single-protein effector complexes) CRISPR-Cas systems. (B) CRISPR-Cas adaptive immunity is mediated by CRISPR RNAs (crRNAs) and Cas proteins, which form multicomponent CRISPR ribonucleoprotein (crRNP) complexes. The first stage is adaptation, which occurs upon entry of an invading mobile genetic element (in this case, a viral genome). Cas1 (blue) and Cas2 (yellow) proteins select and process the invading DNA, and thereafter, a protospacer (orange) is integrated as a new spacer at the leader end of the CRISPR array [repeat sequences (gray) that separate similar-sized, invader-derived spacers (multiple colors)]. During the second stage, expression, the CRISPR locus is transcribed and the pre-crRNA is processed into mature crRNA guides by Cas (e.g., Cas6) or non-Cas proteins (e.g., RNase III). During the final interference stage, the Cas-crRNA complex scans invading DNA for a complementary nucleic acid target, after which the target is degraded by a Cas nuclease. From](/wiki/images/thumb/f/f2/F1r.jpg/450px-F1r.jpg)

