User:Camille Zumstein/Sandbox
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
'''Discovery of the structure:''' | '''Discovery of the structure:''' | ||
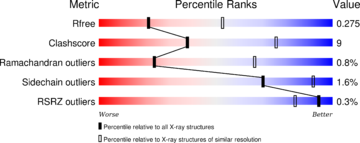
| - | The structure have been published in the year 2013 by Qilu Ye et al. <ref> | + | The structure have been published in the year 2013 by Qilu Ye et al. <ref>PMID:24018048</ref>. For their experiments they used [https://en.wikipedia.org/wiki/X-ray_crystallography#X-ray_diffraction x-ray diffraction]. The PDB validation obtained a Resolutionof 3.0 Å, a free R-value of 0.273 and a work R-value of 0.241 <ref>http://www.rcsb.org/pdb/explore/explore.do?structureId=4IL1</ref>. |
[[Image:4il1 multipercentile validation.png|thumb|upright=2|left]] | [[Image:4il1 multipercentile validation.png|thumb|upright=2|left]] | ||
Revision as of 17:59, 8 January 2017
Structure Rat Calcineurin
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ http://www.jimmunol.org/content/177/4/2681.full
- ↑ http://www.uniprot.org/uniprot/P63329
- ↑ http://www.uniprot.org/uniprot/P63329
- ↑ Ye Q, Feng Y, Yin Y, Faucher F, Currie MA, Rahman MN, Jin J, Li S, Wei Q, Jia Z. Structural basis of calcineurin activation by calmodulin. Cell Signal. 2013 Sep 7;25(12):2661-2667. doi: 10.1016/j.cellsig.2013.08.033. PMID:24018048 doi:10.1016/j.cellsig.2013.08.033
- ↑ http://www.rcsb.org/pdb/explore/explore.do?structureId=4IL1
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/22654726
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/8811062
- ↑ http://www.uptodate.com/contents/pharmacology-of-cyclosporine-and-tacrolimus
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/8837775
- ↑ Calmodulin and Signal Transduction (p184), Linda J. Van Eldik,D. Martin Watterson (1998)
- ↑ http://www.uptodate.com/contents/pharmacology-of-cyclosporine-and-tacrolimus
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/12851457
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/16988714
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2857609/
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/8837775