|
|
| (33 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| | ==Neutrophil Serine 4 or Serine Protease 57 (4Q7X)== | | ==Neutrophil Serine 4 or Serine Protease 57 (4Q7X)== |
| | <StructureSection load='4Q7X' size='350' side='right' caption='[[4Q7X]], [[Resolution|resolution]] 2.55Å' scene=''> | | <StructureSection load='4Q7X' size='350' side='right' caption='[[4Q7X]], [[Resolution|resolution]] 2.55Å' scene=''> |
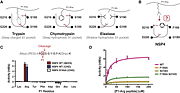
| - | NSP4 also called Neutrophil Serine 4 (NSP4) or Serine Protease 57 (PRSS57),is member of the peptidase S1 family of proteins. It is one of the neutrophil serine protease in the cytoplasmic granules of neutrophils. This enzyme is expressed in bone marrow and takes part of the immune response against pathogens. NSP4 is located in the telomeric region of the chromosome 19. The protein cleaves preferentially after Arg residues but also after citrulline and methylarginie residues. | + | NSP4 also called Neutrophil Serine 4 (NSP4) or Serine Protease 57 (PRSS57), is a member of the peptidase S1 family of proteins. It is one of the neutrophil serine protease in the cytoplasmic granules of neutrophils. This enzyme is expressed in bone marrow and takes part of the immune response against pathogens. NSP4 is located in the telomeric region of the chromosome 19. The protein cleaves preferentially after Arg residues but also after citrulline and methylarginie residues. |
| | | | |
| | | | |
| | ==Function== | | ==Function== |
| | | | |
| - | Recruitement of neutrophils to the site of inflammation is the earliest defense reaction against pathogens. NSP4 is stored in the azurophilic granules [https://www.ncbi.nlm.nih.gov/pubmed/23904161] which is a compartment of neutrophils. In response to neutrophil activation, NSP4 is released into the pericellular environment where microbes are after their phagocytosis. The serine protease 54 not only kill pathogens but also regulate the activity of immune mediators such as chemokines and lymphocytes. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3341072/] NSP4 alterates chemokines by proteolitically cleaving N-terminus of the chemokine.[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016231/] Furthermore, the caspase-like activity of PRSS57 can activate lymphocytes and the adaptive immune response. However, although NSP4 can inactivate some inflammatory processes, the neutrophil serine protease generally promotes than inhibits the inflammatory response. | + | Recruitement of neutrophils to the site of inflammation is the earliest defense reaction against pathogens. NSP4 is stored in the azurophilic granules <ref>PMID:23904161</ref> which is a compartment of neutrophils. In response to neutrophil activation, NSP4 is released into the pericellular environment where microbes are after their phagocytosis. The serine protease 54 doesn't only kill pathogens but also regulates the activity of immune mediators such as chemokines and lymphocytes. <ref>NSP4, an elastase-related protease in human neutrophils with arginine specificity |
| | + | Natascha C. Perera,a Oliver Schilling,b Heike Kittel,a Walter Back,c Elisabeth Kremmer,d and Dieter E. Jenne https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3341072/</ref> NSP4 alterates chemokines by proteolitically cleaving N-terminus of the chemokine.<ref>ailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response |
| | + | Kai Kessenbrock,corresponding author1 Therese Dau,2 and Dieter E. Jenne2 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016231/</ref> Furthermore, the caspase-like activity of PRSS57 can activate lymphocytes and the adaptive immune response. However, although NSP4 can inactivate some inflammatory processes, the neutrophil serine protease generally promotes rather than inhibits the inflammatory response. |
| | | | |
| | | | |
| | == Structural highlights == | | == Structural highlights == |
| | | | |
| - | The protein has a 283 amino-acids long sequence with two mainly domains [[Image:Linear structure of PRSS57.png | thumb]]. It forms a kind of elongate sphere of approximately 70Å large and 150Å long (dimensions: 70,37Å X 70,37Å X 105,02Å). The protein is composed of signal peptide (1-31) which leads to the location in azurophil granules [http://www.jimmunol.org/content/191/5/2700] and allows its excretion. In addition there is a protease domain (32-283) which is a trypsin-like domain with a trypsin-like <scene name='75/751133/Active_site/1'>active site</scene> , according to the specificity for P1-Arg residues, but this domain can be an elastase-like active site according to the primary sequence (because of the presence of a swallow S1 pocket) specific to small aliphatic residues.<ref>S.. Jack Lin, Ken C. Dong, Charles Eigenbrot, Menno van Lookeren Campagne, Daniel Kirchhofer Structures of Neutrophil Serine Protease 4 Reveal an Unusual Mechanism of Substrate Recognition by a Trypsin-Fold Protease DOI: http://dx.doi.org/10.1016/j.str.2014.07.008</ref> | + | The protein has a 283 amino-acids long sequence with two mainly domains [[Image:Linear structure of PRSS57.png | thumb]]. It forms a kind of elongate sphere of approximately 70Å large and 150Å long (dimensions: 70,37Å X 70,37Å X 105,02Å). The protein is composed of signal peptide (1-31) which leads to the location in azurophilic granules <ref>NSP4 Is Stored in Azurophil Granules and Released by Activated Neutrophils as Active Endoprotease with Restricted Specificity |
| - | The <scene name='75/751133/Active_site/1'>active site</scene> is form by 4 amino acids: Gly(189), Phe(190, Ser(216), D(226) | + | Natascha C. Perera, Karl-Heinz Wiesmüller, Maria Torp Larsen, Beate Schacher, Peter Eickholz, Niels Borregaard and Dieter E. Jenne DOI: https://doi.org/10.4049/jimmunol.1301293 </ref> and allows its excretion. In addition there is a protease domain (32-283) which is a trypsin-like domain with a trypsin-like <scene name='75/751133/Active_site/1'>active site</scene> , according to the specificity for P1-Arg residues, but this domain can be an elastase-like active site according to the primary sequence (because of the presence of a swallow S1 pocket) specific to small aliphatic residues.<ref>S.. Jack Lin, Ken C. Dong, Charles Eigenbrot, Menno van Lookeren Campagne, Daniel Kirchhofer Structures of Neutrophil Serine Protease 4 Reveal an Unusual Mechanism of Substrate Recognition by a Trypsin-Fold Protease DOI: http://dx.doi.org/10.1016/j.str.2014.07.008</ref> |
| - | The residue F190 obstructs the active site which could normally not links a P1-Arg. However, a study <ref>Natascha C. Perera, Karl-Heinz Wiesmüller, Maria Torp Larsen, Beate Schacher, Peter Eickholz, Niels Borregaard and Dieter E. Jenne NSP4 Is Stored in Azurophil Granules and Released by Activated Neutrophils as Active Endoprotease with Restricted Specificity DOI: https://doi.org/10.4049/jimmunol.1301293</ref> considered the possibility that the two residues S216 and F190 of the active site can form a flexible gate which allows P1-Arg to enter. Then, the link between the active site and P1-Arg can be stabilized by a salt bridge interaction between S1-D226 and P1-Arg. | + | The <scene name='75/751133/Active_site/1'>active site</scene> is form by 4 amino acids: Gly(189), Phe(190), Ser(216), D(226) |
| - | The hypothesis of the flexible gate was confirmed by the same study. The mutations of S216 only, F190 only or both together show a forced full open gate is more efficient than a forced partially open gate. | + | The residue F190 obstructs the active site which normally couldn't link a P1-Arg. However, a study <ref>Natascha C. Perera, Karl-Heinz Wiesmüller, Maria Torp Larsen, Beate Schacher, Peter Eickholz, Niels Borregaard and Dieter E. Jenne NSP4 Is Stored in Azurophil Granules and Released by Activated Neutrophils as Active Endoprotease with Restricted Specificity DOI: https://doi.org/10.4049/jimmunol.1301293</ref> considers the possibility that the two residues S216 and F190 of the active site can form a flexible gate which allows P1-Arg to enter. Then, the link between the active site and P1-Arg can be stabilized by a salt bridge interaction between S1-D226 and P1-Arg. |
| | + | The hypothesis of the flexible gate is confirmed by the same study. The mutations of S216 only, F190 only or both together show a forced full open gate is more efficient than a forced partially open gate. |
| | The conclusion is that the protease domain is a trypsin-like domain.[[Image:Active site.jpg | thumb]] | | The conclusion is that the protease domain is a trypsin-like domain.[[Image:Active site.jpg | thumb]] |
| | | | |
| Line 20: |
Line 23: |
| | == Interaction == | | == Interaction == |
| | | | |
| - | Like in other trypsin-like proteases D226 is inaccessible to the substrate but helps stabilize the closed S1 pocket by forming an H-bond with the F190 amide. The 99-loop which forms the S2 and S4 substrate interaction sites, is weak and may be implicated in NSP4 allosteric regulation.
| + | As in other trypsin-like proteases, <scene name='75/751133/F190/1'>D226</scene> is inaccessible to the substrate but helps stabilize the closed S1 pocket by forming an H-bond with the <scene name='75/751133/F190/1'>F190</scene> amide. |
| - | NSP4 compared to the other trypsin-like proteases has some specificity as the arginine side chain movement from the canonical "down" to the noncanonical "up" position in NSP4, which is accomplished by a rotation of the Chi2 angle by 160°, this is possible thanks to F190 which provides a hydrophobic platform that interacts with the aliphatic portion of the P1-arginine side chain. All other residue positions are preserved. The specificity for P1-arginine is conferred by H-bonds between guanidinium group and three H-bonds acceptors (S216, S192, G217). This specificity allows NSP4 to cleave citrulline, which is not cleaved by other trypsin-like proteases because of its non-charged propriety, and the impossibility to do salt-bridges. Other trypsin-like proteases are not able to cleave methylarginine neither because of a steric clash with the methyl group, which is not an issue for NSP4 since the methyl group is exposed to the solvent. | + | NSP4 compared to the other trypsin-like proteases has some specificity as the arginine side chain movement from the canonical "down" to the noncanonical "up" position in NSP4, which is accomplished by a rotation of the Chi2 angle by 160°. This is possible thanks to <scene name='75/751133/F190/1'>F190</scene> which provides a hydrophobic platform that interacts with the aliphatic portion of the P1-arginine side chain. All other residue positions are preserved. The specificity for P1-arginine is given by H-bonds between guanidinium group and <scene name='75/751133/H_bounds/1'>three H-bonds acceptors (S216, S192, G217)</scene>. This specificity allows NSP4 to cleave citrulline which is not cleaved by other trypsin-like proteases, because of its non-charged propriety and the impossibility to do salt-bridges. Other trypsin-like proteases are not able to cleave methylarginine because of a steric clash with the methyl group. This is not an issue for NSP4 since the methyl group is exposed to the solvent. The capacity to cleave this post-translationally modified arginine utility may be to act against microbial and virulence factors containing modified arginine or to interact with chemokines. <ref>Lin, S. Jack, Ken C. Dong, Charles Eigenbrot, Menno van Lookeren Campagne, and Daniel Kirchhofer. “Structures of Neutrophil Serine Protease 4 Reveal an Unusual Mechanism of Substrate Recognition by a Trypsin-Fold Protease.” Structure 22, no. 9 (September 2, 2014): 1333–40.[http://dx.doi.org/10.1016/j.str.2014.07.008]</ref> |
| - | The capacity to cleave these post-translationally modified arginine utility may be to act against microbial and virulence factors containing modified arginine or to interact with chemokines. | + | |
| | | | |
| - | N terminus of NSP4 has been shown to be cleaved off by Cathepsin C and occurs during the translocation into the endoplasmic reticulum before the enzyme is stored in cytoplasmic granules. Natural serine protease inhibitors of NSP4 are present in human plasma and can form covalent NSP4-serpin complexes in vitro (antithrombin for example which act as a suicide substrate with NSP4). However, in vivo, antithrombin can not trap NSP4 because of the presence of other neutrophil proteases. | + | <scene name='75/751133/Signal_peptide/1'>N terminus of NSP4</scene> has been shown to be cleaved off by Cathepsin C and occurs during the translocation into the endoplasmic reticulum before the enzyme is stored in cytoplasmic granules. Natural serine protease inhibitors of NSP4 are present in human plasma and can form covalent NSP4-serpin complexes in vitro (for example antithrombin which acts as a suicide substrate with NSP4). However, in vivo, antithrombin can not trap NSP4 because of the presence of other neutrophil proteases.<ref>Perera, Natascha C., Karl-Heinz Wiesmüller, Maria Torp Larsen, Beate Schacher, Peter Eickholz, Niels Borregaard, and Dieter E. Jenne. “NSP4 Is Stored in Azurophil Granules and Released by Activated Neutrophils as Active Endoprotease with Restricted Specificity.” The Journal of Immunology 191, no. 5 (September 1, 2013) [https://doi.org/10.4049/jimmunol.1301293]</ref> |
| | | | |
| | | | |
| | == Disease == | | == Disease == |
| | | | |
| - | Granulocytes neutrophils are the most abundant leukocytes in the blood and are implicated in the immune system. They are the first defend barriers in the innate immunity. When a pathogen agent enters into the body, they recognize it thanks to its antigens and phagocyte the pathogen. They are quick and their recognition is non-specific, and not dependent on previous exposure to microorganisms.[[https://www.ncbi.nlm.nih.gov/pubmed/20836000]] It has also been shown that neutrophils have not only a phagocyte role but can influence all the innate and adaptive immune response by exchanging information with the other immune cells through soluble mediators or direct cell contact.[[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4783580/]]
| + | Neutrophils are the most abundant leukocytes in the blood and are implicated in the immune system. They are the first defend barriers in the innate immunity. When a pathogen agent enters into the body, they recognize it thanks to its antigens and phagocyte the pathogen. They are quick, their recognition is non-specific and not dependent on previous exposure to microorganisms.<ref>Role of neutrophils in innate immunity: a systems biology-level approach. |
| - | In this manner, deficiency in NSP4 might lead to neutrophil deficiency and have terrible consequences. Unfortunately, NP4 is not the more important serin protease of neutrophils, so there isn’t currently in vivo studies of the release of the protein, and any disease as yet been associated with a NSP4 dysfunction. | + | Kobayashi SD1, DeLeo FR. https://www.ncbi.nlm.nih.gov/pubmed/20836000</ref> It has also been shown that neutrophils have not only a phagocyte role but can influence all the innate and adaptive immune response by exchanging information with the other immune cells through soluble mediators or direct cell contact.<ref>Neutrophils: Their Role in Innate and Adaptive Immunity |
| - | Nevertheless, many studies have been released on the three most important neutrophil serin protease: neutrophil elastase (NE), cathepsin G (CG), and proteinase 3 (PR3). As they have quite the same function in the neutrophil, we can hypothesize that the diseases found related to those proteins are related to the diseases we might found with NSP4. | + | Carlos Rosales, 1 , * Nicolas Demaurex, 2 Clifford A. Lowell, 3 and Eileen Uribe-Querol 4 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4783580/</ref> |
| - | For example, instead of phagocyte bacteria, neutrophils can begin an alternative cell death program leading the exchange of their nuclear chromatin by extracellular chromatin fibers covered with antimicrobial protease that can kill bacteria. [[https://www.ncbi.nlm.nih.gov/pubmed/15001782]] However, if neutrophils are over activated it could lead to autoimmune diseases against neutrophils serine proteases or hypersensitivity reactions. | + | In this way, deficiency in NSP4 might leads to neutrophils deficiency and has terrible consequences. Unfortunately, NP4 is not the more important serin protease of neutrophils, so there isn’t currently in vivo studies of the release of the protein. Any disease has yet been associated with a NSP4 dysfunction. |
| - | A disorder of neutrophil serine proteases as also been demonstrated in many genetic disorders like Higashi syndrome or Papillon-Lefevre syndrome.[[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016231/#CR2]] | + | Nevertheless, many studies have been released on the three most important neutrophil serin proteases : neutrophil elastase (NE), cathepsin G (CG) and proteinase 3 (PR3). As they have quite the same function in the neutrophil, we can hypothesize that the diseases found related to those proteins are related to the diseases we might found with NSP4. |
| - | </StructureSection> | + | For example, instead of phagocytising bacteria, neutrophils can begin an alternative cell death program leading the exchange of their nuclear chromatin by extracellular chromatin fibers covered with antimicrobial protease that can kill bacteria. <ref>Neutrophil extracellular traps kill bacteria. |
| - | | + | Brinkmann V1, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. https://www.ncbi.nlm.nih.gov/pubmed/15001782</ref> However, if neutrophils are over activated, it could lead to autoimmune diseases against neutrophils serine proteases or hypersensitivity reactions. |
| - | | + | A disorder of neutrophil serine proteases has also been demonstrated in many genetic disorders like Higashi syndrome or Papillon-Lefevre syndrome.<ref>Tailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response |
| - | | + | Kai Kessenbrock,corresponding author1 Therese Dau,2 and Dieter E. Jenne2 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016231/#CR2</ref> |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| | | | |
| | | | |
| Line 48: |
Line 45: |
| | == References == | | == References == |
| | | | |
| - | [https://www.ncbi.nlm.nih.gov/pubmed/23904161] NSP4 is stored in azurophil granules and released by activated neutrophils as active endoprotease with restricted specificity, NCBI | |
| - | | |
| - | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3341072/] NSP4, an elastase-related protease in human neutrophils with arginine specificity, NCBI | |
| - | | |
| - | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016231/] Tailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response, NCBI | |
| - | | |
| - | [https://www.ncbi.nlm.nih.gov/pubmed/20836000] Role of neutrophils in innate immunity: a systems biology-level approach, NCBI | |
| - | | |
| - | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4783580/] Neutrophils: Their Role in Innate and Adaptive Immunity, NCBI | |
| - | | |
| - | [https://www.ncbi.nlm.nih.gov/pubmed/15001782] Neutrophil extracellular traps kill bacteria, NCBI | |
| - | | |
| - | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016231/#CR2] Tailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response, NCBI | |
| | | | |
| | <references/> | | <references/> |
| NSP4 also called Neutrophil Serine 4 (NSP4) or Serine Protease 57 (PRSS57), is a member of the peptidase S1 family of proteins. It is one of the neutrophil serine protease in the cytoplasmic granules of neutrophils. This enzyme is expressed in bone marrow and takes part of the immune response against pathogens. NSP4 is located in the telomeric region of the chromosome 19. The protein cleaves preferentially after Arg residues but also after citrulline and methylarginie residues.
Function
Recruitement of neutrophils to the site of inflammation is the earliest defense reaction against pathogens. NSP4 is stored in the azurophilic granules [1] which is a compartment of neutrophils. In response to neutrophil activation, NSP4 is released into the pericellular environment where microbes are after their phagocytosis. The serine protease 54 doesn't only kill pathogens but also regulates the activity of immune mediators such as chemokines and lymphocytes. [2] NSP4 alterates chemokines by proteolitically cleaving N-terminus of the chemokine.[3] Furthermore, the caspase-like activity of PRSS57 can activate lymphocytes and the adaptive immune response. However, although NSP4 can inactivate some inflammatory processes, the neutrophil serine protease generally promotes rather than inhibits the inflammatory response.
Structural highlights
The protein has a 283 amino-acids long sequence with two mainly domains . It forms a kind of elongate sphere of approximately 70Å large and 150Å long (dimensions: 70,37Å X 70,37Å X 105,02Å). The protein is composed of signal peptide (1-31) which leads to the location in azurophilic granules [4] and allows its excretion. In addition there is a protease domain (32-283) which is a trypsin-like domain with a trypsin-like , according to the specificity for P1-Arg residues, but this domain can be an elastase-like active site according to the primary sequence (because of the presence of a swallow S1 pocket) specific to small aliphatic residues. [5]
The is form by 4 amino acids: Gly(189), Phe(190), Ser(216), D(226)
The residue F190 obstructs the active site which normally couldn't link a P1-Arg. However, a study [6] considers the possibility that the two residues S216 and F190 of the active site can form a flexible gate which allows P1-Arg to enter. Then, the link between the active site and P1-Arg can be stabilized by a salt bridge interaction between S1-D226 and P1-Arg.
The hypothesis of the flexible gate is confirmed by the same study. The mutations of S216 only, F190 only or both together show a forced full open gate is more efficient than a forced partially open gate.
The conclusion is that the protease domain is a trypsin-like domain.
Interaction
As in other trypsin-like proteases, is inaccessible to the substrate but helps stabilize the closed S1 pocket by forming an H-bond with the amide.
NSP4 compared to the other trypsin-like proteases has some specificity as the arginine side chain movement from the canonical "down" to the noncanonical "up" position in NSP4, which is accomplished by a rotation of the Chi2 angle by 160°. This is possible thanks to which provides a hydrophobic platform that interacts with the aliphatic portion of the P1-arginine side chain. All other residue positions are preserved. The specificity for P1-arginine is given by H-bonds between guanidinium group and . This specificity allows NSP4 to cleave citrulline which is not cleaved by other trypsin-like proteases, because of its non-charged propriety and the impossibility to do salt-bridges. Other trypsin-like proteases are not able to cleave methylarginine because of a steric clash with the methyl group. This is not an issue for NSP4 since the methyl group is exposed to the solvent. The capacity to cleave this post-translationally modified arginine utility may be to act against microbial and virulence factors containing modified arginine or to interact with chemokines. [7]
has been shown to be cleaved off by Cathepsin C and occurs during the translocation into the endoplasmic reticulum before the enzyme is stored in cytoplasmic granules. Natural serine protease inhibitors of NSP4 are present in human plasma and can form covalent NSP4-serpin complexes in vitro (for example antithrombin which acts as a suicide substrate with NSP4). However, in vivo, antithrombin can not trap NSP4 because of the presence of other neutrophil proteases.[8]
Disease
Neutrophils are the most abundant leukocytes in the blood and are implicated in the immune system. They are the first defend barriers in the innate immunity. When a pathogen agent enters into the body, they recognize it thanks to its antigens and phagocyte the pathogen. They are quick, their recognition is non-specific and not dependent on previous exposure to microorganisms.[9] It has also been shown that neutrophils have not only a phagocyte role but can influence all the innate and adaptive immune response by exchanging information with the other immune cells through soluble mediators or direct cell contact.[10]
In this way, deficiency in NSP4 might leads to neutrophils deficiency and has terrible consequences. Unfortunately, NP4 is not the more important serin protease of neutrophils, so there isn’t currently in vivo studies of the release of the protein. Any disease has yet been associated with a NSP4 dysfunction.
Nevertheless, many studies have been released on the three most important neutrophil serin proteases : neutrophil elastase (NE), cathepsin G (CG) and proteinase 3 (PR3). As they have quite the same function in the neutrophil, we can hypothesize that the diseases found related to those proteins are related to the diseases we might found with NSP4.
For example, instead of phagocytising bacteria, neutrophils can begin an alternative cell death program leading the exchange of their nuclear chromatin by extracellular chromatin fibers covered with antimicrobial protease that can kill bacteria. [11] However, if neutrophils are over activated, it could lead to autoimmune diseases against neutrophils serine proteases or hypersensitivity reactions.
A disorder of neutrophil serine proteases has also been demonstrated in many genetic disorders like Higashi syndrome or Papillon-Lefevre syndrome.[12]
References
- ↑ Perera NC, Wiesmuller KH, Larsen MT, Schacher B, Eickholz P, Borregaard N, Jenne DE. NSP4 is stored in azurophil granules and released by activated neutrophils as active endoprotease with restricted specificity. J Immunol. 2013 Sep 1;191(5):2700-7. doi: 10.4049/jimmunol.1301293. Epub 2013 Jul, 31. PMID:23904161 doi:http://dx.doi.org/10.4049/jimmunol.1301293
- ↑ NSP4, an elastase-related protease in human neutrophils with arginine specificity
Natascha C. Perera,a Oliver Schilling,b Heike Kittel,a Walter Back,c Elisabeth Kremmer,d and Dieter E. Jenne https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3341072/
- ↑ ailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response
Kai Kessenbrock,corresponding author1 Therese Dau,2 and Dieter E. Jenne2 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016231/
- ↑ NSP4 Is Stored in Azurophil Granules and Released by Activated Neutrophils as Active Endoprotease with Restricted Specificity
Natascha C. Perera, Karl-Heinz Wiesmüller, Maria Torp Larsen, Beate Schacher, Peter Eickholz, Niels Borregaard and Dieter E. Jenne DOI: https://doi.org/10.4049/jimmunol.1301293
- ↑ S.. Jack Lin, Ken C. Dong, Charles Eigenbrot, Menno van Lookeren Campagne, Daniel Kirchhofer Structures of Neutrophil Serine Protease 4 Reveal an Unusual Mechanism of Substrate Recognition by a Trypsin-Fold Protease DOI: http://dx.doi.org/10.1016/j.str.2014.07.008
- ↑ Natascha C. Perera, Karl-Heinz Wiesmüller, Maria Torp Larsen, Beate Schacher, Peter Eickholz, Niels Borregaard and Dieter E. Jenne NSP4 Is Stored in Azurophil Granules and Released by Activated Neutrophils as Active Endoprotease with Restricted Specificity DOI: https://doi.org/10.4049/jimmunol.1301293
- ↑ Lin, S. Jack, Ken C. Dong, Charles Eigenbrot, Menno van Lookeren Campagne, and Daniel Kirchhofer. “Structures of Neutrophil Serine Protease 4 Reveal an Unusual Mechanism of Substrate Recognition by a Trypsin-Fold Protease.” Structure 22, no. 9 (September 2, 2014): 1333–40.[1]
- ↑ Perera, Natascha C., Karl-Heinz Wiesmüller, Maria Torp Larsen, Beate Schacher, Peter Eickholz, Niels Borregaard, and Dieter E. Jenne. “NSP4 Is Stored in Azurophil Granules and Released by Activated Neutrophils as Active Endoprotease with Restricted Specificity.” The Journal of Immunology 191, no. 5 (September 1, 2013) [2]
- ↑ Role of neutrophils in innate immunity: a systems biology-level approach.
Kobayashi SD1, DeLeo FR. https://www.ncbi.nlm.nih.gov/pubmed/20836000
- ↑ Neutrophils: Their Role in Innate and Adaptive Immunity
Carlos Rosales, 1 , * Nicolas Demaurex, 2 Clifford A. Lowell, 3 and Eileen Uribe-Querol 4 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4783580/
- ↑ Neutrophil extracellular traps kill bacteria.
Brinkmann V1, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. https://www.ncbi.nlm.nih.gov/pubmed/15001782
- ↑ Tailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response
Kai Kessenbrock,corresponding author1 Therese Dau,2 and Dieter E. Jenne2 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3016231/#CR2
|