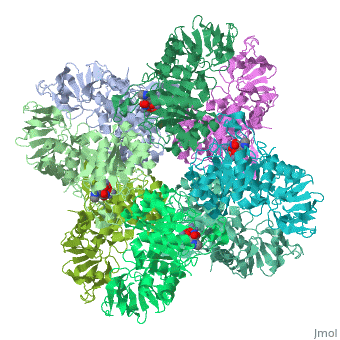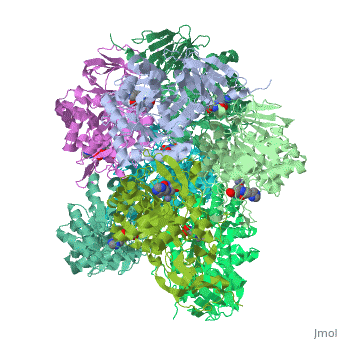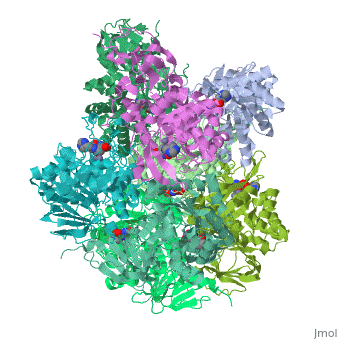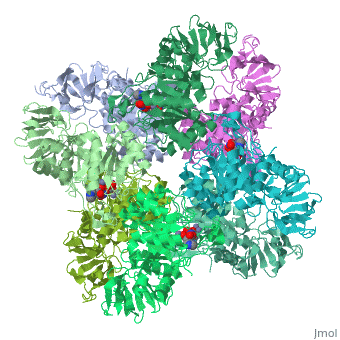
Isoaspartyl dipeptidase
From Proteopedia
(Difference between revisions)
(New page: <StructureSection load='1ybq' size='340' side='right' caption='Caption for this structure' scene=''> == Function == '''Isoaspartyl dipeptidase''' (IadA) catalyzes the hydrolytic cleavage...) |
|||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1ybq' size='340' side='right' caption='E. coli isoaspartyl dipeptidase complex with aspartylhistidine and Zn+2 ions (PDB code [[1ybq]])' scene=''> | |
| - | <StructureSection load='1ybq' size='340' side='right' caption=' | + | |
== Function == | == Function == | ||
| Line 7: | Line 6: | ||
== Structural highlights == | == Structural highlights == | ||
| + | The active site of aspartyl dipeptidase contains the substrate and a binuclear metal center which activates the nucleophilic water molecule<ref>PMID:15882050</ref>. | ||
Revision as of 11:07, 31 January 2017
| |||||||||||
3D structures of dehalogenase
Updated on 31-January-2017
1po9, 1pok, 1onw – EcIadA – Escherichia coli
2aqo, 2aqv – EcIadA (mutant)
1ybq – EcIadA (mutant) + beta-aspartylhistidine
1poj – EcIadA + inhibitor
1onx – EcIadA + aspartate
References
- ↑ Michalska K, Brzezinski K, Jaskolski M. Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate. J Biol Chem. 2005 Aug 5;280(31):28484-91. Epub 2005 Jun 9. PMID:15946951 doi:10.1074/jbc.M504501200
- ↑ Marti-Arbona R, Fresquet V, Thoden JB, Davis ML, Holden HM, Raushel FM. Mechanism of the reaction catalyzed by isoaspartyl dipeptidase from Escherichia coli. Biochemistry. 2005 May 17;44(19):7115-24. PMID:15882050 doi:10.1021/bi050008r