1v3v
From Proteopedia
| Line 4: | Line 4: | ||
|PDB= 1v3v |SIZE=350|CAPTION= <scene name='initialview01'>1v3v</scene>, resolution 2.0Å | |PDB= 1v3v |SIZE=350|CAPTION= <scene name='initialview01'>1v3v</scene>, resolution 2.0Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= <scene name='pdbligand= | + | |LIGAND= <scene name='pdbligand=5OP:(5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC+ACID'>5OP</scene>, <scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=NAP:NADP+NICOTINAMIDE-ADENINE-DINUCLEOTIDE+PHOSPHATE'>NAP</scene> |
| - | |ACTIVITY= [http://en.wikipedia.org/wiki/15-oxoprostaglandin_13-oxidase 15-oxoprostaglandin 13-oxidase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.3.1.48 1.3.1.48] | + | |ACTIVITY= <span class='plainlinks'>[http://en.wikipedia.org/wiki/15-oxoprostaglandin_13-oxidase 15-oxoprostaglandin 13-oxidase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.3.1.48 1.3.1.48] </span> |
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY=[[1v3t|1V3T]], [[1v3u|1V3U]] | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1v3v FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1v3v OCA], [http://www.ebi.ac.uk/pdbsum/1v3v PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1v3v RCSB]</span> | ||
}} | }} | ||
| Line 33: | Line 36: | ||
[[Category: Yamamoto, M.]] | [[Category: Yamamoto, M.]] | ||
[[Category: Yokomizo, T.]] | [[Category: Yokomizo, T.]] | ||
| - | [[Category: 5OP]] | ||
| - | [[Category: CL]] | ||
| - | [[Category: NAP]] | ||
[[Category: riken structural genomics/proteomics initiative]] | [[Category: riken structural genomics/proteomics initiative]] | ||
[[Category: rossmann fold]] | [[Category: rossmann fold]] | ||
| Line 41: | Line 41: | ||
[[Category: structural genomic]] | [[Category: structural genomic]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 00:19:07 2008'' |
Revision as of 21:19, 30 March 2008
| |||||||
| , resolution 2.0Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||
| Activity: | 15-oxoprostaglandin 13-oxidase, with EC number 1.3.1.48 | ||||||
| Related: | 1V3T, 1V3U
| ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
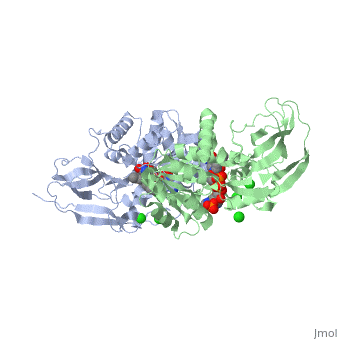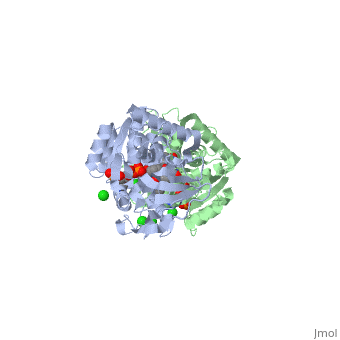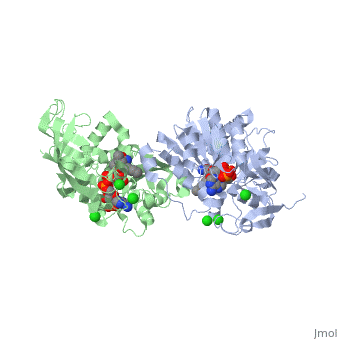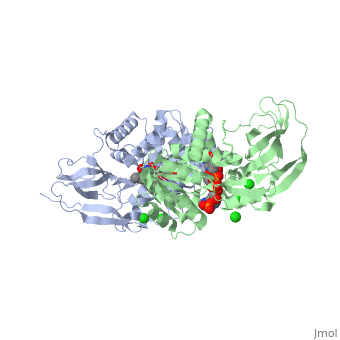
Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complexed with NADP and 15-oxo-PGE2
Overview
The bifunctional leukotriene B(4) 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase (LTB(4) 12-HD/PGR) is an essential enzyme for eicosanoid inactivation. It is involved in the metabolism of the E and F series of 15-oxo-prostaglandins (15-oxo-PGs), leukotriene B(4) (LTB(4)), and 15-oxo-lipoxin A(4) (15-oxo-LXA(4)). Some nonsteroidal anti-inflammatory drugs (NSAIDs), which primarily act as cyclooxygenase inhibitors also inhibit LTB(4) 12-HD/PGR activity. Here we report the crystal structure of the LTB(4) 12-HD/PGR, the binary complex structure with NADP(+), and the ternary complex structure with NADP(+) and 15-oxo-PGE(2). In the ternary complex, both in the crystalline form and in solution, the enolate anion intermediate accumulates as a brown chromophore. PGE(2) contains two chains, but only the omega-chain of 15-oxo-PGE(2) was defined in the electron density map in the ternary complex structure. The omega-chain was identified at the hydrophobic pore on the dimer interface. The structure showed that the 15-oxo group forms hydrogen bonds with the 2'-hydroxyl group of nicotine amide ribose of NADP(+) and a bound water molecule to stabilize the enolate intermediate during the reductase reaction. The electron-deficient C13 atom of the conjugated enolate may be directly attacked by a hydride from the NADPH nicotine amide in a stereospecific manner. The moderate recognition of 15-oxo-PGE(2) is consistent with a broad substrate specificity of LTB(4) 12-HD/PGR. The structure also implies that a Src homology domain 3 may interact with the left-handed proline-rich helix at the dimer interface and regulate LTB(4) 12-HD/PGR activity by disruption of the substrate binding pore to accommodate the omega-chain.
About this Structure
1V3V is a Single protein structure of sequence from Cavia porcellus. Full crystallographic information is available from OCA.
Reference
Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop., Hori T, Yokomizo T, Ago H, Sugahara M, Ueno G, Yamamoto M, Kumasaka T, Shimizu T, Miyano M, J Biol Chem. 2004 May 21;279(21):22615-23. Epub 2004 Mar 8. PMID:15007077
Page seeded by OCA on Mon Mar 31 00:19:07 2008
Categories: 15-oxoprostaglandin 13-oxidase | Cavia porcellus | Single protein | Ago, H. | Hori, T. | Kumasaka, T. | Miyano, M. | RSGI, RIKEN Structural Genomics/Proteomics Initiative. | Shimizu, T. | Sugahara, M. | Ueno, G. | Yamamoto, M. | Yokomizo, T. | Riken structural genomics/proteomics initiative | Rossmann fold | Rsgi | Structural genomic