UDP-N-acetylglucosamine acyltransferase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
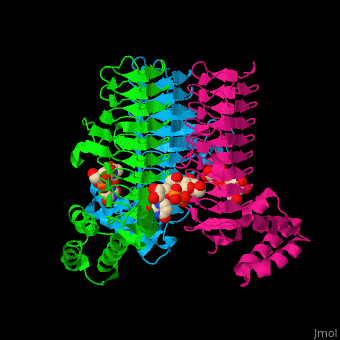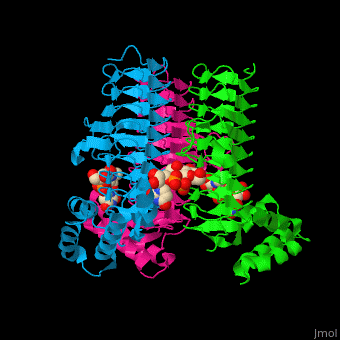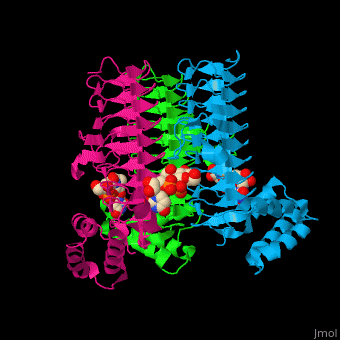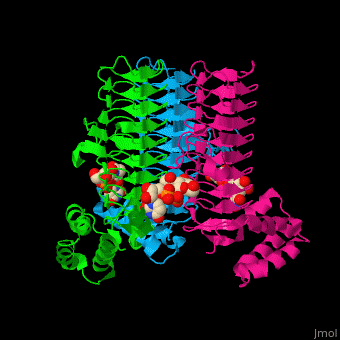
| - | <StructureSection load='2jf3' size=' | + | <StructureSection load='2jf3' size='450' side='right' caption='E. coli UDP-N-acetylglucosamine acyltransferase complex with UDP-Glc-NAC (PDB code [[2jf3]])' scene='74/749395/Cv/1'> |
== Function == | == Function == | ||
Revision as of 12:49, 8 June 2017
| |||||||||||
3D structures of UDP-N-acetylglucosamine acyltransferase
Updated on 08-June-2017
References
- ↑ Wyckoff TJ, Raetz CR. The active site of Escherichia coli UDP-N-acetylglucosamine acyltransferase. Chemical modification and site-directed mutagenesis. J Biol Chem. 1999 Sep 17;274(38):27047-55. PMID:10480918
- ↑ Ulaganathan V, Buetow L, Hunter WN. Nucleotide substrate recognition by UDP-N-acetylglucosamine acyltransferase (LpxA) in the first step of lipid A biosynthesis. J Mol Biol. 2007 Jun 1;369(2):305-12. Epub 2007 Mar 21. PMID:17434525 doi:10.1016/j.jmb.2007.03.039