Aldehyde dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
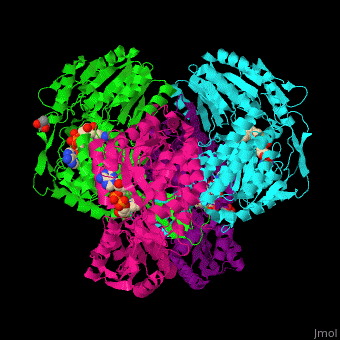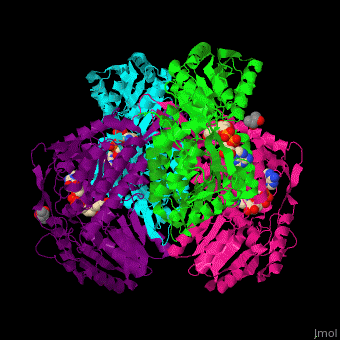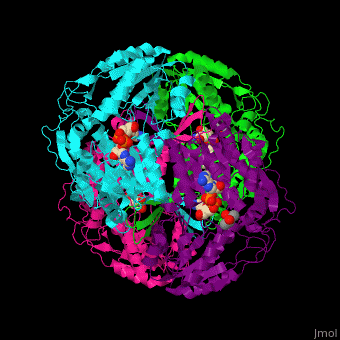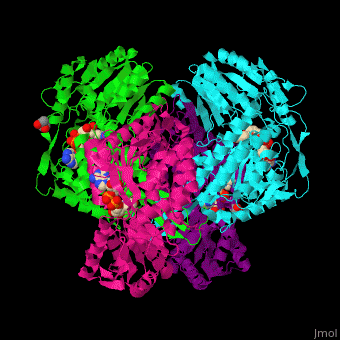
| - | <StructureSection load='1o9j' size=' | + | <StructureSection load='1o9j' size='450' side='right' scene='44/447036/Cv/2' caption='Aldehyde hydrogenase class 1 tetramer complex with NAD, dithiothreitol and dimercaptobutane-diol, [[1o9j]]'> |
== Function == | == Function == | ||
| Line 20: | Line 20: | ||
== Structural highlights == | == Structural highlights == | ||
| - | A cysteine-thiol at the active site of GAPDH plays a role in catalysis. <scene name='44/447036/Cv/ | + | A cysteine-thiol at the active site of GAPDH plays a role in catalysis. |
| + | *<scene name='44/447036/Cv/5'>Active site</scene>. | ||
</StructureSection> | </StructureSection> | ||
== 3D Structures of Aldehyde dehydrogenase == | == 3D Structures of Aldehyde dehydrogenase == | ||
Revision as of 09:11, 7 August 2017
| |||||||||||
3D Structures of Aldehyde dehydrogenase
Updated on 07-August-2017
References
- ↑ Keller, Markus A.; Zander, Ulrich; Fuchs, Julian E.; Kreutz, Christoph; Watschinger, Katrin et al. (2014). A gatekeeper helix determines the substrate specificity of Sjögren–Larsson Syndrome enzyme fatty aldehyde dehydrogenase. Nature Communications vol. 5.