1xbn
From Proteopedia
| Line 4: | Line 4: | ||
|PDB= 1xbn |SIZE=350|CAPTION= <scene name='initialview01'>1xbn</scene>, resolution 2.50Å | |PDB= 1xbn |SIZE=350|CAPTION= <scene name='initialview01'>1xbn</scene>, resolution 2.50Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= <scene name='pdbligand= | + | |LIGAND= <scene name='pdbligand=HEM:PROTOPORPHYRIN+IX+CONTAINING+FE'>HEM</scene>, <scene name='pdbligand=OXY:OXYGEN+MOLECULE'>OXY</scene> |
|ACTIVITY= | |ACTIVITY= | ||
|GENE= Tar4 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=119072 Thermoanaerobacter tengcongensis]) | |GENE= Tar4 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=119072 Thermoanaerobacter tengcongensis]) | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1xbn FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1xbn OCA], [http://www.ebi.ac.uk/pdbsum/1xbn PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1xbn RCSB]</span> | ||
}} | }} | ||
| Line 24: | Line 27: | ||
[[Category: Nioche, P.]] | [[Category: Nioche, P.]] | ||
[[Category: Raman, C S.]] | [[Category: Raman, C S.]] | ||
| - | [[Category: HEM]] | ||
| - | [[Category: OXY]] | ||
[[Category: cgmp]] | [[Category: cgmp]] | ||
[[Category: heme protein]] | [[Category: heme protein]] | ||
| Line 31: | Line 32: | ||
[[Category: soluble guanylyl cyclase]] | [[Category: soluble guanylyl cyclase]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 00:47:24 2008'' |
Revision as of 21:47, 30 March 2008
| |||||||
| , resolution 2.50Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Gene: | Tar4 (Thermoanaerobacter tengcongensis) | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
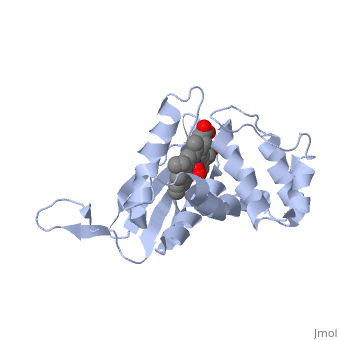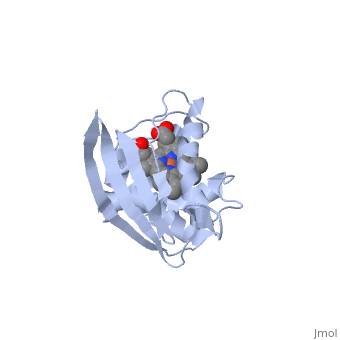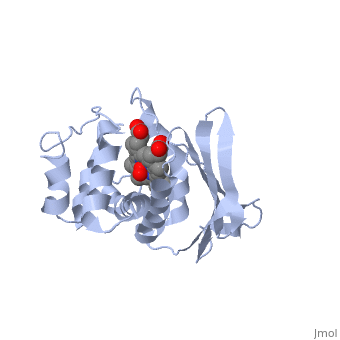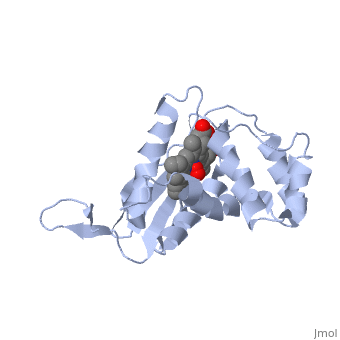
Crystal structure of a bacterial nitric oxide sensor: an ortholog of mammalian soluble guanylate cyclase heme domain
Overview
Nitric oxide (NO) is extremely toxic to Clostridium botulinum, but its molecular targets are unknown. Here, we identify a heme protein sensor (SONO) that displays femtomolar affinity for NO. The crystal structure of the SONO heme domain reveals a previously undescribed fold and a strategically placed tyrosine residue that modulates heme-nitrosyl coordination. Furthermore, the domain architecture of a SONO ortholog cloned from Chlamydomonas reinhardtii indicates that NO signaling through cyclic guanosine monophosphate arose before the origin of multicellular eukaryotes. Our findings have broad implications for understanding bacterial responses to NO, as well as for the activation of mammalian NO-sensitive guanylyl cyclase.
About this Structure
1XBN is a Single protein structure of sequence from Thermoanaerobacter tengcongensis. Full crystallographic information is available from OCA.
Reference
Femtomolar sensitivity of a NO sensor from Clostridium botulinum., Nioche P, Berka V, Vipond J, Minton N, Tsai AL, Raman CS, Science. 2004 Nov 26;306(5701):1550-3. Epub 2004 Oct 7. PMID:15472039
Page seeded by OCA on Mon Mar 31 00:47:24 2008