This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Journal:Structure:1
From Proteopedia
(Difference between revisions)

| Line 6: | Line 6: | ||
Protein-protein interactions (PPI) mediate most major processes in the cell. Despite the crowded cellular environment, proteins maintain a high degree of specificity in their interactions. For this, proteins evolved to balance between the ability to bind the desired partners while rejecting all other proteins. | Protein-protein interactions (PPI) mediate most major processes in the cell. Despite the crowded cellular environment, proteins maintain a high degree of specificity in their interactions. For this, proteins evolved to balance between the ability to bind the desired partners while rejecting all other proteins. | ||
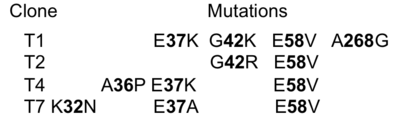
| - | Here, we address the question of the sequence distance to generate new binding. In other words, how many mutations have to be inserted into a protein so that it will bind a given partner. For this we generated a random library of TEM1-b-lactamase mutant proteins displayed on yeast and selected the library to bind wild-type TEM1. We found that three mutations were sufficient to develop a de-novo protein interaction (<scene name='76/763767/Cv/2'>Figure 1</scene> and Table below). | + | Here, we address the question of the sequence distance to generate new binding. In other words, how many mutations have to be inserted into a protein so that it will bind a given partner. For this we generated a random library of TEM1-b-lactamase mutant proteins displayed on yeast and selected the library to bind wild-type TEM1. We found that three mutations were sufficient to develop a de-novo protein interaction (<scene name='76/763767/Cv/2'>Figure 1</scene> and Table below). The position of each of the selected mutations is represented as orange sticks on the TEM1-WT scaffold (PDB [[1jtg]] chain A). |
{{Clear}} | {{Clear}} | ||
[[Image:Gid1.png|thumb|The identities of the mutations present in the 4 selected clones are provided in the table|400px|left]] | [[Image:Gid1.png|thumb|The identities of the mutations present in the 4 selected clones are provided in the table|400px|left]] | ||
Revision as of 10:19, 18 October 2017
| |||||||||||
- ↑ REF
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.