Journal:Structure:1
From Proteopedia
(Difference between revisions)

| Line 12: | Line 12: | ||
The mutations severely destabilized the protein, making them accessible only on a pre-stabilized TEM1 variant<ref>pmid 23542341 </ref>. The X-ray structure of the complex formed between the mutant TEM1 (termed T1 G268A) and TEM1 wild-type showed the two proteins to be related by a <scene name='76/763767/Cv/10'>pseudo-two-fold symmetry axis</scene>. The interface is comprised of strand 2 from both proteins forming a <scene name='76/763767/Cv/11'>continuous β-sheet</scene>, which spans to strand 1 and forms a <scene name='76/763767/Cv/13'>perfect backbone hydrogen bond network</scene>. The most surprising feature of the complex is that helix 1 of T1 G268A is completely missing in the density map. <scene name='76/763767/Cv/18'>Overlaying TEM1-WT onto T1 G268A</scene> shows that a properly folded N'-helix in T1 G268A would physically interfere with the interaction, making complex formation impossible. The T1 G268 interface on TEM1-WT does not overlay the BLIP binding site ([[1jtg]]), allowing for both to bind simultaneously. | The mutations severely destabilized the protein, making them accessible only on a pre-stabilized TEM1 variant<ref>pmid 23542341 </ref>. The X-ray structure of the complex formed between the mutant TEM1 (termed T1 G268A) and TEM1 wild-type showed the two proteins to be related by a <scene name='76/763767/Cv/10'>pseudo-two-fold symmetry axis</scene>. The interface is comprised of strand 2 from both proteins forming a <scene name='76/763767/Cv/11'>continuous β-sheet</scene>, which spans to strand 1 and forms a <scene name='76/763767/Cv/13'>perfect backbone hydrogen bond network</scene>. The most surprising feature of the complex is that helix 1 of T1 G268A is completely missing in the density map. <scene name='76/763767/Cv/18'>Overlaying TEM1-WT onto T1 G268A</scene> shows that a properly folded N'-helix in T1 G268A would physically interfere with the interaction, making complex formation impossible. The T1 G268 interface on TEM1-WT does not overlay the BLIP binding site ([[1jtg]]), allowing for both to bind simultaneously. | ||
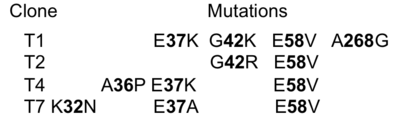
| - | A closer look at the three mutations evolved during selection shows that two of them (E37K and E58V) are important in stabilizing the N'-helix in its position. Analysis of the <scene name='76/763767/Cv/20'>WT structure</scene> ([[1btl]]) shows that <scene name='76/763767/Cv/21'>E37 forms an intra-protein salt bridge with R61</scene>, which in turn <scene name='76/763767/Cv/22'>interacts with E64</scene>, which <scene name='76/763767/Cv/23'>interacts with R43</scene>. <scene name='76/763767/Cv/ | + | A closer look at the three mutations evolved during selection shows that two of them (E37K and E58V) are important in stabilizing the N'-helix in its position. Analysis of the <scene name='76/763767/Cv/20'>WT structure</scene> ([[1btl]]) shows that <scene name='76/763767/Cv/21'>E37 forms an intra-protein salt bridge with R61</scene>, which in turn <scene name='76/763767/Cv/22'>interacts with E64</scene>, which <scene name='76/763767/Cv/23'>interacts with R43</scene>. <scene name='76/763767/Cv/27'>E58 interacts with the NH2-terminus of TEM1-WT</scene>. Therefore, loosing E37 and E58 will destabilize the N'- helix, as indeed seen in the <scene name='76/763767/Cv/10'>T1 structure</scene>. The two additional selected mutations, K32 and A36 in some of the clones probably also contribute to destabilization of the N'helix. The only selected mutation located directly in the binding interface is E58V, which is present in all four selected clones. E58V forms hydrophobic contacts with L30 and F60 of the other chain. The structure implies that the formed interaction would be very specific between the evolved mutants and TEM1-WT. The structure of the complex shows that one copy of the N'-helix is important in stabilizing the interaction between the two proteins. Hence the WT cannot dimerize since the N'-helix physically interferes with the interaction while the selected TEM1 proteins are missing a folded N'-helix. A detailed map of the interactions formed between the two proteins is presented in the figure below. |
{{Clear}} | {{Clear}} | ||
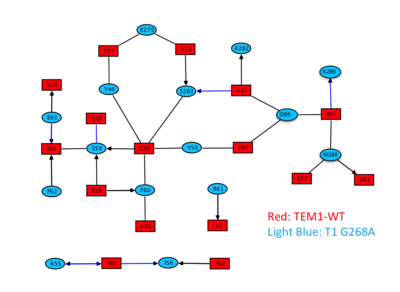
[[Image:Slide4Gid.png|thumb|Interacting residues from both proteins as predicted by AquaPort<ref>pmid 17070843 </ref> are presented in red (TEM1-WT) and light blue (T1 G268A). The chemical bond formed between the interacting residues is indicated by the type of their connecting line, black: hydrophobic; blue: polar; line: interaction via side chain; arrow: interaction via main chain. |400px|left]] | [[Image:Slide4Gid.png|thumb|Interacting residues from both proteins as predicted by AquaPort<ref>pmid 17070843 </ref> are presented in red (TEM1-WT) and light blue (T1 G268A). The chemical bond formed between the interacting residues is indicated by the type of their connecting line, black: hydrophobic; blue: polar; line: interaction via side chain; arrow: interaction via main chain. |400px|left]] | ||
Revision as of 07:55, 24 October 2017
| |||||||||||
- ↑ REF
- ↑ Dellus-Gur E, Toth-Petroczy A, Elias M, Tawfik DS. What Makes a Protein Fold Amenable to Functional Innovation? Fold Polarity and Stability Trade-offs. J Mol Biol. 2013 Mar 28. pii: S0022-2836(13)00200-3. doi:, 10.1016/j.jmb.2013.03.033. PMID:23542341 doi:10.1016/j.jmb.2013.03.033
- ↑ Reichmann D, Cohen M, Abramovich R, Dym O, Lim D, Strynadka NC, Schreiber G. Binding hot spots in the TEM1-BLIP interface in light of its modular architecture. J Mol Biol. 2007 Jan 19;365(3):663-79. Epub 2006 Oct 3. PMID:17070843 doi:10.1016/j.jmb.2006.09.076
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.