Sandbox Reserved 1382
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
Actin provides unique advantages that make this solution more dependable, flexible, and scalable than alternatives. Actin is self-propelled, operates independently, and has small dimensions which makes it able to explore the dense grid network in a parallel manner. ATP is used to power the computation, which eliminates the need of an electric potential to be delivered from a single point of access, which makes powering larger SSP computations realistic. The molecular motor attached to the F-actin is also much more power efficient than traditional computers, effectively eliminating heat dissipation limitations.<ref>doi:10.1073/pnas.1510825113</ref> The actin design can be mass produced as a computing agent for all NP-complete problems since the nature of the problem is inherently encoded into the grid network. Furthermore, actin can also replenish itself through enzymatic splitting and elongation. | Actin provides unique advantages that make this solution more dependable, flexible, and scalable than alternatives. Actin is self-propelled, operates independently, and has small dimensions which makes it able to explore the dense grid network in a parallel manner. ATP is used to power the computation, which eliminates the need of an electric potential to be delivered from a single point of access, which makes powering larger SSP computations realistic. The molecular motor attached to the F-actin is also much more power efficient than traditional computers, effectively eliminating heat dissipation limitations.<ref>doi:10.1073/pnas.1510825113</ref> The actin design can be mass produced as a computing agent for all NP-complete problems since the nature of the problem is inherently encoded into the grid network. Furthermore, actin can also replenish itself through enzymatic splitting and elongation. | ||
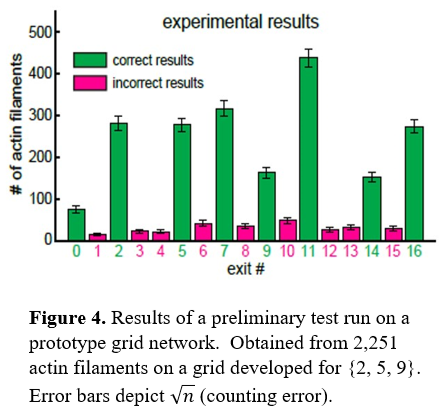
| - | [[Image:Figure 4..png|frame|center| | + | [[Image:Figure 4..png|frame|center|100px]] |
The preliminary test run on a 3 number SSP provides error rate too high to be within a reasonable margin of error for SSPs with more than 10 numbers (Fig. 4). The error is a direct result of the failure of pass junctions to force 100% of F-actin to traverse in a straight path.<ref>doi:10.1073/pnas.1510825113</ref> Still, the success rate for SSPs with less than 10 variables is acceptable enough to be a viable method of parallel computation. | The preliminary test run on a 3 number SSP provides error rate too high to be within a reasonable margin of error for SSPs with more than 10 numbers (Fig. 4). The error is a direct result of the failure of pass junctions to force 100% of F-actin to traverse in a straight path.<ref>doi:10.1073/pnas.1510825113</ref> Still, the success rate for SSPs with less than 10 variables is acceptable enough to be a viable method of parallel computation. | ||
Revision as of 21:41, 22 February 2018
| This Sandbox is Reserved from January through July 31, 2018 for use in the course HLSC322: Principles of Genetics and Genomics taught by Genevieve Houston-Ludlam at the University of Maryland, College Park, USA. This reservation includes Sandbox Reserved 1311 through Sandbox Reserved 1430. |
To get started:
More help: Help:Editing |
Globular Actin
| |||||||||||
References
- ↑ doi: https://dx.doi.org/10.2210/rcsb_pdb/mom_2001_7
- ↑ Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169-86. doi: 10.1146/annurev-biophys-042910-155359. PMID:21314430 doi:http://dx.doi.org/10.1146/annurev-biophys-042910-155359
- ↑ Nicolau DV Jr, Lard M, Korten T, van Delft FC, Persson M, Bengtsson E, Mansson A, Diez S, Linke H, Nicolau DV. Parallel computation with molecular-motor-propelled agents in nanofabricated networks. Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2591-6. doi:, 10.1073/pnas.1510825113. Epub 2016 Feb 22. PMID:26903637 doi:http://dx.doi.org/10.1073/pnas.1510825113
- ↑ Nicolau DV Jr, Lard M, Korten T, van Delft FC, Persson M, Bengtsson E, Mansson A, Diez S, Linke H, Nicolau DV. Parallel computation with molecular-motor-propelled agents in nanofabricated networks. Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2591-6. doi:, 10.1073/pnas.1510825113. Epub 2016 Feb 22. PMID:26903637 doi:http://dx.doi.org/10.1073/pnas.1510825113