User:Andrea Foote/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Structure == | == Structure == | ||
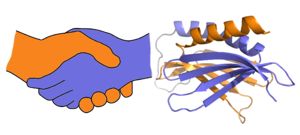
| - | <scene name='78/786627/5fgo_repeatiii/2'>PUR repeat III</scene> facilitates dimerization | + | Purα functions as a dimer composed of two intramolecular domains and one intermolecular domain. The Purα monomer contains three semi-conserved repeated amino acid sequences, named in order from N->C: PUR repeats I, II, and III. These repeats fold to form two domains: <scene name='78/786627/5fgp_intro/8'>PUR repeats I and II</scene> associating to form the I-II domain or “intramolecular domain”, while <scene name='78/786627/5fgo_repeatiii/2'>PUR repeat III</scene> facilitates dimerization through association with a repeat III from a second Purα monomer or repeat III of Purβ. Each PUR repeat is connected by flexible linker regions. |
| - | [[Image:180429 proteopedia pura figures2.jpg|thumb|right|300px| A PUR domain is analogous to a left-handed handshake. PUR repeat I-II represented from 5fgp. | + | [[Image:180429 proteopedia pura figures2.jpg|thumb|right|300px| A PUR domain is analogous to a left-handed handshake. PUR repeat I-II represented from 5fgp.]] |
| - | ]] | + | |
== Function == | == Function == | ||
| - | + | High-affinity nucleic acid-binding function is dependent on Purα dimerization. Purα forms homodimers in addition to heterodimers with Purβ. PurA is known to repress various genes including | |
<scene name='78/786627/5fgp_57and145/1'>Two aromatic residues</scene>, Y57 (repeat I) and F145 (repeat II) have been implicated in the DNA unwinding activity of PurA.<ref>PMID:26744780</ref> | <scene name='78/786627/5fgp_57and145/1'>Two aromatic residues</scene>, Y57 (repeat I) and F145 (repeat II) have been implicated in the DNA unwinding activity of PurA.<ref>PMID:26744780</ref> | ||
== Disease == | == Disease == | ||
| - | == Relevance == | ||
| - | |||
| - | == Structural highlights == | ||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> | ||
| - | </StructureSection> | ||
| - | == Function == | ||
| - | <StructureSection load='5fgp' size='340' side='right' caption= scene='78/786627/5fgp_57and145/1'> | ||
| - | |||
| - | Anything in this section will appear adjacent to the 3D structure and will be scrollable. | ||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 10:46, 3 May 2018
Purine-rich element binding protein alpha
| |||||||||||