|
|
| (233 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | [[User: Julie Langlois/PsaA]] | + | =='''Alginate Binding Periplasmin Proteins of Sphingomonas sp. A1 (AlgQ1, AlgQ2)'''== |
| | + | <StructureSection load='1y3n' size='350' side='right' caption='Structure of AlgQ1, alginate-binding protein, complexed with an alginate disaccharide (PDB entry [[1y3n]])' scene=''> |
| | | | |
| | | | |
| - | NCBI Accession: P42363.1
| + | == Introduction: == |
| - | Uniprot Accesion: POA4G2
| + | |
| - | PDB ID: 3ZK7
| + | |
| | | | |
| - | <StructureSection load='3zk7' size='340' side='right' caption='[[3zk7]], [[Resolution|resolution]] 1.69Å' scene=''> | + | Alginate binding protein is the protein responsible for mediating the transport of alginate from the pit on the cell surface to the alginate specific ABC (ATP-binding cassette) importer <ref>PMID:15794643</ref>. This system of transferring exists in a gram-negative bacterium, Sphingomonas sp. A1<ref>PMID:8785434</ref>. This protein, which is a periplasmic binding protein, has two homologues AlgQ1 and AlgQ2 coded in PDB as 1Y3N and 1J1N respectively. Alginate is an anionic polysaccharide that is highly found in the cell walls of brown algae. Alginic acid (Alginate) is a linear copolymer with homopolymeric blocks of (1-4)-linked β-D-mannuronate (M) and its C-5 epimer α-L-guluronate (G) residues, linked with covalent bonds. Monomers arrange together in three forms, blocks of consecutive G residues, Blocks of Consecutive M residues and heteropolymeric random sequences of G and M<ref>PMID:22125349</ref>. Strain A1 directly take in this polymeric molecule into the cytoplasm in a process, an important part of which is alginate-binding proteins. 1Y3N is the structure of this alginate binding protein complexed with an alginate disaccharide. A significant number of ABC transporters analyzed so far are just capable of transporting small molecules with a molecular mass less than 2 kDa <ref>PMID:15189142</ref>. In macromolecule assimilation, the macromolecule degrading enzymes play the role of making smaller molecules by breaking macromolecules into pieces, so the living cell can assimilate it. As a result, the alginate ABC importer of strain A1 is unusual in the sense that it can import a macromolecule with an average molecular mass of 26 kDa, without the need to break the macromolecule into smaller parts and the alginate binding protein is the periplasmic binding protein that mediates this transport along with other proteins being active in this specific transporter <ref>PMID:9529892</ref>. |
| | + | This structure has <scene name='55/559112/1y3n_ligands/2'>three bound ligands</scene>, <scene name='55/559112/Bem_494_beta-d-mannuronic_acid/1'>BEM</scene>, <scene name='55/559112/Mav_495_alpha-d-mannopyranuron/1'>MAV</scene> and Calcium atom. BEM is beta-D-mannuronic acid, and MAV is alpha-D-mannopyranuronic acid. The two-dimensional structure of these two ligands is provided here.[[image:2dim.jpg|thumb|left|alt=BEM (494) and MAV (495) two-dimensional structures]] |
| | | | |
| - | == Overview == | |
| - | === PsaA protein === | |
| - | PsaA (Pneumococcal surface antigen A) is a multi-functional lipoprotein detected on all known serotypes of ''Streptococcus pneumoniae''. This lipoprotein is part of the ABC-type (ATP binding complex) transport protein complex that transports Mn2+. PsaA is also an adhesion factor that plays a major role in pneumococcal attachment to the host cell and virulence. PsaA is hidden beneath the cell wall. PsaA protein is involved in in colonization of the nasopharyngeal mucosal. | |
| - | This transporter is composed of the products of three genes, psaB (ATP-binding protein), psaC (integral membrane protein), and psaA (solute-binding lipoprotein), which are organized in an operon with a gene encoding PsaD, a thiol peroxidase <ref>PMID:517531</ref>. | |
| | | | |
| - | [[Image:operon psa.jpg |500px| thumb| Psa operon <ref>PMID:15255900</ref>]]
| + | ---- |
| - |
| + | |
| - | [[Image:Linear structure PsaA.jpg |500px| thumb|Linear structure of PsaA]]
| + | |
| | | | |
| | + | [[Image:ABC Transporter.jpg|thumb|'This is a summary of how Strain A1 superchannel works to import and degrade alginate. G, L-guluronate; M, D-mannuronate; gene for alginate lyases (A1-I, A1-II and A1-III); catabolite-control protein gene; algS, algM1 and algM2, ABC transporter genes for alginate import; algQ1 and algQ2, genes for alginate-binding proteins; a1-IV, alginate lyase A1-IV gene; a1-II’, alginate lyase A1-II’ gene; a1-IV’, alginate lyase A1-IV’ gene; p3, TonB-dependent transporter; p5, alginate receptor; FlgJ, C-terminal catalytic module for peptidoglycan hydrolysis. Adopted from Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi:http://dx.doi.org/10.4161/bbug.1.2.10322']] |
| | | | |
| | + | ---- |
| | | | |
| | | | |
| | + | == Structural Description and insights to function: == |
| | | | |
| | | | |
| | + | The alginate binding protein consists of two main groups <scene name='55/559112/Algq1/1'>AlgQ1</scene> and <scene name='55/559112/Algq2/1'>AlgQ2</scene>. AlgQ1 and AlgQ2 are two periplasmic proteins with the almost very similar function which is mediating the transport of the substrate and studies show that the primary structure for this two is about 76% similar, although further investigations are yet to be done to clarify the function and structure differences between these two<ref>PMID:15794643</ref>.Here, we just cover the general structure and functions of these two AlgQ1 and AlgQ2 and their binding to carbohydrates (Alginate). The function of a protein is determined by its shape and the shape of a protein is determined by its primary structure (sequence of amino acids). Based on the analysis of the primary structure done by ([http://www.psort.org/]) and ProtScale ([http://kr.expasy.org/tools/ protscale.html]), this protein is water soluble secretory, which suggests that it is periplasmic<ref>PMID:15794643</ref>. The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria<ref>PMID:29342145</ref>. The N-terminal sequence of AlgQ1 is REATW (Arginine, Glutamic acid, Alanine, Threonine, Tryptophan). The first 24 amino acid residues play the role of a <scene name='55/559112/Signal_peptide/1'>signal peptide</scene>. A signal peptide is a short peptide including between 16 to 30 amino acids, which is found at the N-terminus of proteins that are destined toward the secretory pathway<ref>Kapp, Katja; Schrempf, Sabrina; Lemberg, Marius K.; Dobberstein, Bernhard (2013-01-01). Post-Targeting Functions of Signal Peptides. Landes Bioscience.</ref>. The bulk of this part includes 5 to 16 hydrophobic amino acids. These hydrophobic residues tend to create a single α-helix and are also referred to as “h-region”. there is typically a number of amino acids at the end of the signal peptide that is recognized and cleaved by signal peptidase and therefore named cleavage site<ref>PMID:15794643</ref>. |
| | + | AlgQ1 in its two forms (Apo and Holo) comprises 490 amino acid residues, indicating that proteins are truncated. The C-terminal amino acid is determined to be Tyrosine. In addition, the amino acid analysis suggests that AlgQ1 is truncated between Tyr490 and Gly491<ref>PMID:15794643</ref>.We can see the apo-AlgQ1 as a cartoon presentation, made up of <scene name='55/559112/Doamins_of_algq1/1'>two globular domains</scene> N domain and c domain, consisting of residues 1-133, 310-400 and 134-309, 401-490 respectively. The two domains that we introduced are connected to each other through 3 loop segments consisting of residues 133-136, 292-347, and 399-401. The disaccharide is bound to the deep cleft between N and C domains. As mentioned in CATH classification, these two domain both are in a class of alpha/beta proteins, showing 3-Layer(aba) Sandwich architecture, having the topology of D-Maltodextrin-Binding Proteins; domain 2 and the homology of both of these domains as can be predicted is Periplasmic binding protein-like II. If we want to assess how SCOP classify this protein, we can see that SCOP sees this protein as a one domain totally, but the same as in CATH, from the alpha and beta proteins. In addition, this protein belongs to Periplasmic binding protein-like superfamily and comes from Phosphate binding protein-like family<ref>PMID:15794643</ref><ref>http://www.cathdb.info/</ref>. |
| | + | When N-domains of apo and holo forms of the protein were superimposed, then a rotation angle of 0.6 degrees was needed for the C domain to be superimposed. Structures of a holo-AlgQ with tetrasaccharide and disaccharide (AlgQ1) are almost the same<ref>PMID:15794643</ref>. |
| | + | Here we can see the binding sites holo-algQ1-DI. This bound consists of ΔM1-M2 with α-anomeric M2 at S1 and S2. ΔM, M, and G denote unsaturated D-mannuronate, saturated D-mannuronate, and saturated L-guluronate, respectively. |
| | + | The bound oligosaccharides interact with surrounding amino acids. As Keiko momma, et al. summarized hydrogen bond interactions between the bound alginate oligosaccharides and alginate binding proteins(data not shown) The number of direct hydrogen bonds between AlgQ1 and the disaccharide in holo-AlgQ1 DI is 11 and the number of associated water molecules is 10. Five water molecules are located at S3 and S4 subsites in holo-AlgQ1-DI. The number of C-C contacts that AlgQ1 and disaccharide holo-AlgQ1-DI have is 30, which indicates that the nonreducing end of sugar is in a significant involvement with AlgQ1<ref>PMID:15794643</ref>. |
| | + | Ramachandran plot analysis shows that most non-glycine residues are located in the most favorable regions. There is only one exception to this, which is Lys251, (apo-AlgQ1, φ=65° and ψ=-141°; holo-AlgQ1-TE, φ=62° and ψ=-133°; and holo-AlgQ1-DI (1Y3N), φ=65° and ψ=-139°), and it is present in an allowed region. Lys251 is located next to the terminus of a helix (H12/C)<ref>PMID:15794643</ref>. |
| | | | |
| | + | The crystal structure of AlgQ2 consist of two domains separated by a cleft and binds and releases alginate tetrasaccharide by creating conformational change in these two domains<ref>PMID:21326935</ref>. To mention some of the different forms of this protein we can take a look at 5H6U, 5H71, 1KWH, 1J1N in PDB. As an alginate binding protein, the dissociation constants are 6 μg/mL for AlgQ1 and 4 μg/mL for AlgQ2.Results from UV absorption spectroscopy indicate that both of these proteins are alginate specific<ref>PMID:15794643</ref>. |
| | | | |
| | | | |
| | + | ---- |
| | | | |
| | + | ==Links to available structures== |
| | + | Alginate binding periplasmic structures with binding to different alginate polymers |
| | | | |
| | + | [http://www.rcsb.org/structure/1Y3N] :1Y3N |
| | | | |
| - | Between the first to the 24th amino acid namely the red motif there is the signal peptide and the second one is the Pfam motif<ref>http://string-db.org/cgi/network.pl?taskId=EA2jsQm5Sc5A</ref>.
| + | [http://www.rcsb.org/structure/1Y3P] :1Y3P |
| | | | |
| | + | [http://www.rcsb.org/structure/1Y3Q] :1Y3Q |
| | | | |
| | + | [http://www.rcsb.org/structure/1KWH] :1KWH |
| | | | |
| | + | [http://www.rcsb.org/structure/1J1N] :1J1N |
| | | | |
| | + | [http://www.rcsb.org/structure/3A09] :3A09 |
| | | | |
| | + | [http://www.rcsb.org/structure/3VLU] :3VLU |
| | | | |
| - | Zinc in excess has significant toxicity to bacteria because it is an important innate defence mechanism. There are many Zinc in human body. Manganese is important for the virulence, growth and proliferation of ''Streptococcus pneumoniae''. Zinc could compete for Manganese binding. However Manganese has more affinity for PsaA than Zinc but Zinc is not transported by the ABC-transporter. Zinc competition reduces intracellular Manganse resulting in up-regulation of PsaBCA expression. <ref>PMID:22072971</ref>
| + | [http://www.rcsb.org/structure/3VLV] :3VLV |
| | | | |
| | + | [http://www.rcsb.org/structure/3VLW]: 3VLW |
| | | | |
| - | [[Image:PsaA dans ABC transporteur.jpg |500px| Structure of ABC-transporter <ref>http://www.latrobe.edu.au/biochemistry-and-genetics/research/maher/psabca-manganese-uptake-by-streptococcus-pneumoniae</ref>]] | + | [http://www.rcsb.org/structure/5H6U] :5H6U |
| | | | |
| | + | [http://www.rcsb.org/structure/5H71] |
| | | | |
| | | | |
| | + | == Link to evolutionary related Structures== |
| | | | |
| | + | By running the protein through Consurf ([http://consurf.tau.ac.il/2016/]), the Phylogenetic tree of 1Y3N can be obtained. Below a link to this tree is provided. |
| | + | [http://consurf.tau.ac.il//wasabi/?url=http://consurf.tau.ac.il/results/1525350845/query_msa_fasta_and_Tree.xml] |
| | | | |
| | + | The <scene name='55/559112/1y3q_evolutionary/1'>evolutionary conservation 3D structure of 1Y3Q (AlgQ1)</scene>is shown here. |
| | | | |
| | | | |
| | + | ---- |
| | | | |
| - | | + | == References: == |
| - | | + | <references /> |
| - | | + | </StructureSection> |
| - | === ''Streptococcus pneumoniae''=== | + | |
| - | ''Streptococcus pneumoniae'' is a Gram positive cocci (with a diameter from 0.5 to 1 μm) and a member of the genus ''Streptococcus''. It can live under aerobic or anaerobic conditions. It resides in the nasopharynx of healthy carriers. However, the bacterium may become pathogenic in elderly and immunocompromised adults and children. Then it can spread to other locations and cause disease. The genome of ''S. pneumoniae'' is a closed, circular DNA structure that contains between 2.0 and 2.1 million base pairs.<ref>PMID:4366526</ref>
| + | |
| - | | + | |
| - | [[Image:streptococcus pneumoniae.jpg | 350px|thumb| ''Streptococcus pneumoniae'' <ref>http://www.sciencephoto.com/images/download_lo_res.html?id=662360183</ref>]]
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | == Structure ==
| + | |
| - | The protein PsaA has a molecular weight of 34.538 kDa with 309 residues<ref>https://www.mybiosource.com/prods/Recombinant-Protein/Manganese-ABC-transporter-substrate-binding-lipoprotein-psaA/psaA</ref>. The overall size of the protein approximated from its crystal structure is 40 by 40 by 70 Å<ref>PMID:28011228</ref>.
| + | |
| - | As a member of the Lipoprotein receptor-associated antigen I (LraI) family, the PsaA molecule contains four distinct regions. An N-terminal leader sequence of 20 amino acids holds an LxACy consensus sequence that is recognized and cleaved by signal peptidase II <ref>PMID:28011228</ref>. A lipid moiety (diacylglycerol <ref>PMID:PMC99024</ref>) is added to the cysteine residue and mediates the anchorage of the protein to the cytoplasmic membrane. Apart from this leader sequence, the rest of the protein consists of two twofold-pseudosymmetrical (β/α)4 sandwich domains, of which the β-strands of each domain form parallel β-sheets <ref>PMID:PMC99024</ref>. In total the two domains form two lobes connected via an α-helical linker which constitutes the solute-binding site <ref>PMID:28011228</ref>.
| + | |
| - | | + | |
| - | <Structure load='3ztt' size='300' color='white' frame='true' align='right' caption='testcaption' />
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | {|
| + | |
| - | | [[Image:details psaa avec mn.jpg]]
| + | |
| - | | [[Image:details psaa avec zn.jpg]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | | + | |
| - | This image shows the metal binding site in more detail, with the MntC residues and Manganese. Manganse ions are shown as purple spheres. We can see that Manganese interacts with Histine residues, Aspartique acid residues and Glutamique acid residues.
| + | |
| - | The metal binding site is formed by the sidechains of residues His67, His139, Glu205, and Asp280. This site has tetrahedral coordination geometry. The amino acids His67 andHis139 interact with the metal via Nε2 nitrogen atoms. However, the carboxylate sidechains of Glu205 and Asp280 interact with the metal via their Oε1 and Oδ2 atoms. The atomic distance between His67 Nε2 and the metal is 1.99 Ắ while it is 2,01 Ắ for His 139 Nε2. It is 2,04 Ắ for Glu205 Oε1 and 2,02 Ắ for Asp280 Oδ2. The Oδ1 atom of Asp137 can form a hydrogen bond with Nδ1 of His139. Oδ1 atom of Asp65 can also form a hydrogen bond with His67 N. <ref>PMID:9862808</ref>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | This image shows the metal binding site in more detail, with MntC residues and Managanese and Zinc. Manganese ion is shown as a purple sphere and Zinc ion is shown as an orange sphere. Cadmium uptake reduces the millimolar cellular accumulation of manganese and zinc, and thereby increases sensitivity to oxidative stress <ref>http://www.nature.com/articles/ncomms7418</ref>.
| + | |
| - | | + | |
| - | [[Image:molec.jpg|left|350px | thumb]]
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | 3D structure of PsaA with 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL (tris) and cadmium<ref>http://www.ebi.ac.uk/pdbe/entry/pdb/4UTO</ref>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | == Protein-protein interaction ==
| + | |
| - | | + | |
| - | == Function ==
| + | |
| - | Mn2+ is also required for the activity of CpsB, a tyrosine phosphatase involved in the regulation of capsule production. And in some streptococcal species, lectin-mediated adherence requires Mn2+. Finally, mutations in the psa operon result in an almost complete attenuation of virulence for all tested models of animal infection.
| + | |
| - | == Disease ==
| + | |
| - | === Meningitis ===
| + | |
| - | === Otitis ===
| + | |
| - | PsaA has been recognized to be involved in the adherence and virulence mechanisms of otitis media. Streptococcus pneumoniae is one of the main agents causing bacterial acute otitis media, directly or as complication of a viral upper respiratory tract infection. This disease is a highly prevalent pediatric disease worldwide. Hearing loss is a common problem associated with this disease. <ref>https://www.google.com/patents/US20130078254</ref>
| + | |
| - | === Pneumonia ===
| + | |
| - | PsaA protein is an adhesin which is involved in colonization of the nasopharyngeal mucosal. Moreover, alveolar pneumonia is caused by the spread of ''Streptococcus pneumoniae'' from nasopharynx. Therefore PsaA protein is involved in infection of pulmonary parenchyma by ''Streptococcus pneumoniae''.
| + | |
| - | Sometimes, ''Streptococcus pneumoniae'' pass into the blood and causes a bacteremia besides pneumonia.
| + | |
| - | == Application in Biotechnology ==
| + | |
| - | PsaA is being actively evaluated as a component of a vaccin in formulations composed of pneumococcal common proteins. PsaA is a component of a vaccin because this protein is immunogenic and stimulates an increase in antibody production when the nasopharynx is naturally colonized. PsaA has been expressed as an ''E.coli'' recombinant protein, purified, and evaluated in a phase one clinical trial.
| + | |
| - | Progress in vaccine development is most advanced for Streptococcus pneumoniae. Indeed, there is a seven-valent capsular-conjugate vaccine, PREVNAR® but it is rather non efficient for otitis media.<ref>PMID:11176564</ref>
| + | |
| - | ==References==
| + | |
| - | <references/>
| + | |
|
Introduction:
Alginate binding protein is the protein responsible for mediating the transport of alginate from the pit on the cell surface to the alginate specific ABC (ATP-binding cassette) importer [1]. This system of transferring exists in a gram-negative bacterium, Sphingomonas sp. A1[2]. This protein, which is a periplasmic binding protein, has two homologues AlgQ1 and AlgQ2 coded in PDB as 1Y3N and 1J1N respectively. Alginate is an anionic polysaccharide that is highly found in the cell walls of brown algae. Alginic acid (Alginate) is a linear copolymer with homopolymeric blocks of (1-4)-linked β-D-mannuronate (M) and its C-5 epimer α-L-guluronate (G) residues, linked with covalent bonds. Monomers arrange together in three forms, blocks of consecutive G residues, Blocks of Consecutive M residues and heteropolymeric random sequences of G and M[3]. Strain A1 directly take in this polymeric molecule into the cytoplasm in a process, an important part of which is alginate-binding proteins. 1Y3N is the structure of this alginate binding protein complexed with an alginate disaccharide. A significant number of ABC transporters analyzed so far are just capable of transporting small molecules with a molecular mass less than 2 kDa [4]. In macromolecule assimilation, the macromolecule degrading enzymes play the role of making smaller molecules by breaking macromolecules into pieces, so the living cell can assimilate it. As a result, the alginate ABC importer of strain A1 is unusual in the sense that it can import a macromolecule with an average molecular mass of 26 kDa, without the need to break the macromolecule into smaller parts and the alginate binding protein is the periplasmic binding protein that mediates this transport along with other proteins being active in this specific transporter [5].
This structure has , , and Calcium atom. BEM is beta-D-mannuronic acid, and MAV is alpha-D-mannopyranuronic acid. The two-dimensional structure of these two ligands is provided here. 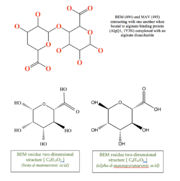 alt=BEM (494) and MAV (495) two-dimensional structures
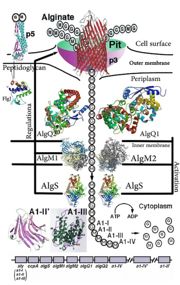 'This is a summary of how Strain A1 superchannel works to import and degrade alginate. G, L-guluronate; M, D-mannuronate; gene for alginate lyases (A1-I, A1-II and A1-III); catabolite-control protein gene; algS, algM1 and algM2, ABC transporter genes for alginate import; algQ1 and algQ2, genes for alginate-binding proteins; a1-IV, alginate lyase A1-IV gene; a1-II’, alginate lyase A1-II’ gene; a1-IV’, alginate lyase A1-IV’ gene; p3, TonB-dependent transporter; p5, alginate receptor; FlgJ, C-terminal catalytic module for peptidoglycan hydrolysis. Adopted from Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi: http://dx.doi.org/10.4161/bbug.1.2.10322'
Structural Description and insights to function:
The alginate binding protein consists of two main groups and . AlgQ1 and AlgQ2 are two periplasmic proteins with the almost very similar function which is mediating the transport of the substrate and studies show that the primary structure for this two is about 76% similar, although further investigations are yet to be done to clarify the function and structure differences between these two[6].Here, we just cover the general structure and functions of these two AlgQ1 and AlgQ2 and their binding to carbohydrates (Alginate). The function of a protein is determined by its shape and the shape of a protein is determined by its primary structure (sequence of amino acids). Based on the analysis of the primary structure done by ([1]) and ProtScale (protscale.html), this protein is water soluble secretory, which suggests that it is periplasmic[7]. The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria[8]. The N-terminal sequence of AlgQ1 is REATW (Arginine, Glutamic acid, Alanine, Threonine, Tryptophan). The first 24 amino acid residues play the role of a . A signal peptide is a short peptide including between 16 to 30 amino acids, which is found at the N-terminus of proteins that are destined toward the secretory pathway[9]. The bulk of this part includes 5 to 16 hydrophobic amino acids. These hydrophobic residues tend to create a single α-helix and are also referred to as “h-region”. there is typically a number of amino acids at the end of the signal peptide that is recognized and cleaved by signal peptidase and therefore named cleavage site[10].
AlgQ1 in its two forms (Apo and Holo) comprises 490 amino acid residues, indicating that proteins are truncated. The C-terminal amino acid is determined to be Tyrosine. In addition, the amino acid analysis suggests that AlgQ1 is truncated between Tyr490 and Gly491[11].We can see the apo-AlgQ1 as a cartoon presentation, made up of N domain and c domain, consisting of residues 1-133, 310-400 and 134-309, 401-490 respectively. The two domains that we introduced are connected to each other through 3 loop segments consisting of residues 133-136, 292-347, and 399-401. The disaccharide is bound to the deep cleft between N and C domains. As mentioned in CATH classification, these two domain both are in a class of alpha/beta proteins, showing 3-Layer(aba) Sandwich architecture, having the topology of D-Maltodextrin-Binding Proteins; domain 2 and the homology of both of these domains as can be predicted is Periplasmic binding protein-like II. If we want to assess how SCOP classify this protein, we can see that SCOP sees this protein as a one domain totally, but the same as in CATH, from the alpha and beta proteins. In addition, this protein belongs to Periplasmic binding protein-like superfamily and comes from Phosphate binding protein-like family[12][13].
When N-domains of apo and holo forms of the protein were superimposed, then a rotation angle of 0.6 degrees was needed for the C domain to be superimposed. Structures of a holo-AlgQ with tetrasaccharide and disaccharide (AlgQ1) are almost the same[14].
Here we can see the binding sites holo-algQ1-DI. This bound consists of ΔM1-M2 with α-anomeric M2 at S1 and S2. ΔM, M, and G denote unsaturated D-mannuronate, saturated D-mannuronate, and saturated L-guluronate, respectively.
The bound oligosaccharides interact with surrounding amino acids. As Keiko momma, et al. summarized hydrogen bond interactions between the bound alginate oligosaccharides and alginate binding proteins(data not shown) The number of direct hydrogen bonds between AlgQ1 and the disaccharide in holo-AlgQ1 DI is 11 and the number of associated water molecules is 10. Five water molecules are located at S3 and S4 subsites in holo-AlgQ1-DI. The number of C-C contacts that AlgQ1 and disaccharide holo-AlgQ1-DI have is 30, which indicates that the nonreducing end of sugar is in a significant involvement with AlgQ1[15].
Ramachandran plot analysis shows that most non-glycine residues are located in the most favorable regions. There is only one exception to this, which is Lys251, (apo-AlgQ1, φ=65° and ψ=-141°; holo-AlgQ1-TE, φ=62° and ψ=-133°; and holo-AlgQ1-DI (1Y3N), φ=65° and ψ=-139°), and it is present in an allowed region. Lys251 is located next to the terminus of a helix (H12/C)[16].
The crystal structure of AlgQ2 consist of two domains separated by a cleft and binds and releases alginate tetrasaccharide by creating conformational change in these two domains[17]. To mention some of the different forms of this protein we can take a look at 5H6U, 5H71, 1KWH, 1J1N in PDB. As an alginate binding protein, the dissociation constants are 6 μg/mL for AlgQ1 and 4 μg/mL for AlgQ2.Results from UV absorption spectroscopy indicate that both of these proteins are alginate specific[18].
Links to available structures
Alginate binding periplasmic structures with binding to different alginate polymers
[2] :1Y3N
[3] :1Y3P
[4] :1Y3Q
[5] :1KWH
[6] :1J1N
[7] :3A09
[8] :3VLU
[9] :3VLV
[10]: 3VLW
[11] :5H6U
[12]
Link to evolutionary related Structures
By running the protein through Consurf ([13]), the Phylogenetic tree of 1Y3N can be obtained. Below a link to this tree is provided.
[14]
The is shown here.
References:
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ White DC, Sutton SD, Ringelberg DB. The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol. 1996 Jun;7(3):301-6. PMID:8785434
- ↑ Lee KY, Mooney DJ. Alginate: properties and biomedical applications. Prog Polym Sci. 2012 Jan;37(1):106-126. doi: 10.1016/j.progpolymsci.2011.06.003. PMID:22125349 doi:http://dx.doi.org/10.1016/j.progpolymsci.2011.06.003
- ↑ Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241-68. doi: 10.1146/annurev.biochem.73.011303.073626. PMID:15189142 doi:http://dx.doi.org/10.1146/annurev.biochem.73.011303.073626
- ↑ Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998 Mar;62(1):204-29. PMID:9529892
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Miller SI, Salama NR. The gram-negative bacterial periplasm: Size matters. PLoS Biol. 2018 Jan 17;16(1):e2004935. doi: 10.1371/journal.pbio.2004935., eCollection 2018 Jan. PMID:29342145 doi:http://dx.doi.org/10.1371/journal.pbio.2004935
- ↑ Kapp, Katja; Schrempf, Sabrina; Lemberg, Marius K.; Dobberstein, Bernhard (2013-01-01). Post-Targeting Functions of Signal Peptides. Landes Bioscience.
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ http://www.cathdb.info/
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi:http://dx.doi.org/10.4161/bbug.1.2.10322
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
|


