Elizeu/sandbox/citocromo c
From Proteopedia
(Difference between revisions)
| (37 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | ==''' | + | =='''Alginate Binding Periplasmin Proteins of Sphingomonas sp. A1 (AlgQ1, AlgQ2)'''== |
| - | <StructureSection load='1y3n' size=' | + | <StructureSection load='1y3n' size='350' side='right' caption='Structure of AlgQ1, alginate-binding protein, complexed with an alginate disaccharide (PDB entry [[1y3n]])' scene=''> |
| + | |||
| + | |||
| + | == Introduction: == | ||
Alginate binding protein is the protein responsible for mediating the transport of alginate from the pit on the cell surface to the alginate specific ABC (ATP-binding cassette) importer <ref>PMID:15794643</ref>. This system of transferring exists in a gram-negative bacterium, Sphingomonas sp. A1<ref>PMID:8785434</ref>. This protein, which is a periplasmic binding protein, has two homologues AlgQ1 and AlgQ2 coded in PDB as 1Y3N and 1J1N respectively. Alginate is an anionic polysaccharide that is highly found in the cell walls of brown algae. Alginic acid (Alginate) is a linear copolymer with homopolymeric blocks of (1-4)-linked β-D-mannuronate (M) and its C-5 epimer α-L-guluronate (G) residues, linked with covalent bonds. Monomers arrange together in three forms, blocks of consecutive G residues, Blocks of Consecutive M residues and heteropolymeric random sequences of G and M<ref>PMID:22125349</ref>. Strain A1 directly take in this polymeric molecule into the cytoplasm in a process, an important part of which is alginate-binding proteins. 1Y3N is the structure of this alginate binding protein complexed with an alginate disaccharide. A significant number of ABC transporters analyzed so far are just capable of transporting small molecules with a molecular mass less than 2 kDa <ref>PMID:15189142</ref>. In macromolecule assimilation, the macromolecule degrading enzymes play the role of making smaller molecules by breaking macromolecules into pieces, so the living cell can assimilate it. As a result, the alginate ABC importer of strain A1 is unusual in the sense that it can import a macromolecule with an average molecular mass of 26 kDa, without the need to break the macromolecule into smaller parts and the alginate binding protein is the periplasmic binding protein that mediates this transport along with other proteins being active in this specific transporter <ref>PMID:9529892</ref>. | Alginate binding protein is the protein responsible for mediating the transport of alginate from the pit on the cell surface to the alginate specific ABC (ATP-binding cassette) importer <ref>PMID:15794643</ref>. This system of transferring exists in a gram-negative bacterium, Sphingomonas sp. A1<ref>PMID:8785434</ref>. This protein, which is a periplasmic binding protein, has two homologues AlgQ1 and AlgQ2 coded in PDB as 1Y3N and 1J1N respectively. Alginate is an anionic polysaccharide that is highly found in the cell walls of brown algae. Alginic acid (Alginate) is a linear copolymer with homopolymeric blocks of (1-4)-linked β-D-mannuronate (M) and its C-5 epimer α-L-guluronate (G) residues, linked with covalent bonds. Monomers arrange together in three forms, blocks of consecutive G residues, Blocks of Consecutive M residues and heteropolymeric random sequences of G and M<ref>PMID:22125349</ref>. Strain A1 directly take in this polymeric molecule into the cytoplasm in a process, an important part of which is alginate-binding proteins. 1Y3N is the structure of this alginate binding protein complexed with an alginate disaccharide. A significant number of ABC transporters analyzed so far are just capable of transporting small molecules with a molecular mass less than 2 kDa <ref>PMID:15189142</ref>. In macromolecule assimilation, the macromolecule degrading enzymes play the role of making smaller molecules by breaking macromolecules into pieces, so the living cell can assimilate it. As a result, the alginate ABC importer of strain A1 is unusual in the sense that it can import a macromolecule with an average molecular mass of 26 kDa, without the need to break the macromolecule into smaller parts and the alginate binding protein is the periplasmic binding protein that mediates this transport along with other proteins being active in this specific transporter <ref>PMID:9529892</ref>. | ||
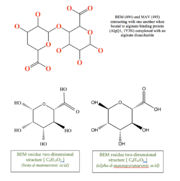
| - | This structure has <scene name='55/559112/1y3n_ligands/2'>three bound ligands</scene>, BEM, MAV and Calcium atom. BEM is beta-D-mannuronic acid, and MAV is alpha-D-mannopyranuronic acid. The two-dimensional structure of these two ligands is provided here. | + | This structure has <scene name='55/559112/1y3n_ligands/2'>three bound ligands</scene>, <scene name='55/559112/Bem_494_beta-d-mannuronic_acid/1'>BEM</scene>, <scene name='55/559112/Mav_495_alpha-d-mannopyranuron/1'>MAV</scene> and Calcium atom. BEM is beta-D-mannuronic acid, and MAV is alpha-D-mannopyranuronic acid. The two-dimensional structure of these two ligands is provided here.[[image:2dim.jpg|thumb|left|alt=BEM (494) and MAV (495) two-dimensional structures]] |
| - | -- | + | ---- |
| + | |||
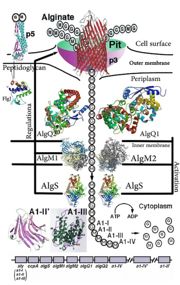
| + | [[Image:ABC Transporter.jpg|thumb|'This is a summary of how Strain A1 superchannel works to import and degrade alginate. G, L-guluronate; M, D-mannuronate; gene for alginate lyases (A1-I, A1-II and A1-III); catabolite-control protein gene; algS, algM1 and algM2, ABC transporter genes for alginate import; algQ1 and algQ2, genes for alginate-binding proteins; a1-IV, alginate lyase A1-IV gene; a1-II’, alginate lyase A1-II’ gene; a1-IV’, alginate lyase A1-IV’ gene; p3, TonB-dependent transporter; p5, alginate receptor; FlgJ, C-terminal catalytic module for peptidoglycan hydrolysis. Adopted from Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi:http://dx.doi.org/10.4161/bbug.1.2.10322']] | ||
| + | |||
| + | ---- | ||
| + | |||
== Structural Description and insights to function: == | == Structural Description and insights to function: == | ||
| Line 15: | Line 23: | ||
When N-domains of apo and holo forms of the protein were superimposed, then a rotation angle of 0.6 degrees was needed for the C domain to be superimposed. Structures of a holo-AlgQ with tetrasaccharide and disaccharide (AlgQ1) are almost the same<ref>PMID:15794643</ref>. | When N-domains of apo and holo forms of the protein were superimposed, then a rotation angle of 0.6 degrees was needed for the C domain to be superimposed. Structures of a holo-AlgQ with tetrasaccharide and disaccharide (AlgQ1) are almost the same<ref>PMID:15794643</ref>. | ||
Here we can see the binding sites holo-algQ1-DI. This bound consists of ΔM1-M2 with α-anomeric M2 at S1 and S2. ΔM, M, and G denote unsaturated D-mannuronate, saturated D-mannuronate, and saturated L-guluronate, respectively. | Here we can see the binding sites holo-algQ1-DI. This bound consists of ΔM1-M2 with α-anomeric M2 at S1 and S2. ΔM, M, and G denote unsaturated D-mannuronate, saturated D-mannuronate, and saturated L-guluronate, respectively. | ||
| - | The bound oligosaccharides interact with surrounding amino acids. As Keiko momma, et al. summarized hydrogen bond interactions between the bound alginate oligosaccharides and alginate binding proteins(data not shown) The number of direct hydrogen bonds between AlgQ1 and the disaccharide in holo-AlgQ1 DI is 11 and the number of associated water molecules is 10. Five water molecules are located at S3 and S4 subsites in holo-AlgQ1-DI. The number of C-C contacts that AlgQ1 and disaccharide holo-AlgQ1-DI have is 30, which indicates that the nonreducing end of sugar is in a significant involvement with AlgQ1. | + | The bound oligosaccharides interact with surrounding amino acids. As Keiko momma, et al. summarized hydrogen bond interactions between the bound alginate oligosaccharides and alginate binding proteins(data not shown) The number of direct hydrogen bonds between AlgQ1 and the disaccharide in holo-AlgQ1 DI is 11 and the number of associated water molecules is 10. Five water molecules are located at S3 and S4 subsites in holo-AlgQ1-DI. The number of C-C contacts that AlgQ1 and disaccharide holo-AlgQ1-DI have is 30, which indicates that the nonreducing end of sugar is in a significant involvement with AlgQ1<ref>PMID:15794643</ref>. |
Ramachandran plot analysis shows that most non-glycine residues are located in the most favorable regions. There is only one exception to this, which is Lys251, (apo-AlgQ1, φ=65° and ψ=-141°; holo-AlgQ1-TE, φ=62° and ψ=-133°; and holo-AlgQ1-DI (1Y3N), φ=65° and ψ=-139°), and it is present in an allowed region. Lys251 is located next to the terminus of a helix (H12/C)<ref>PMID:15794643</ref>. | Ramachandran plot analysis shows that most non-glycine residues are located in the most favorable regions. There is only one exception to this, which is Lys251, (apo-AlgQ1, φ=65° and ψ=-141°; holo-AlgQ1-TE, φ=62° and ψ=-133°; and holo-AlgQ1-DI (1Y3N), φ=65° and ψ=-139°), and it is present in an allowed region. Lys251 is located next to the terminus of a helix (H12/C)<ref>PMID:15794643</ref>. | ||
| - | The crystal structure of AlgQ2 consist of two domains separated by a cleft and binds and releases alginate tetrasaccharide by creating conformational change in these two domains. To mention some of the different forms of this protein we can take a look at 5H6U, 5H71, 1KWH, 1J1N in PDB. | + | The crystal structure of AlgQ2 consist of two domains separated by a cleft and binds and releases alginate tetrasaccharide by creating conformational change in these two domains<ref>PMID:21326935</ref>. To mention some of the different forms of this protein we can take a look at 5H6U, 5H71, 1KWH, 1J1N in PDB. As an alginate binding protein, the dissociation constants are 6 μg/mL for AlgQ1 and 4 μg/mL for AlgQ2.Results from UV absorption spectroscopy indicate that both of these proteins are alginate specific<ref>PMID:15794643</ref>. |
| - | As an alginate binding protein, the dissociation constants are 6 μg/mL for AlgQ1 and 4 μg/mL for AlgQ2. | + | |
| - | Results from UV absorption spectroscopy indicate that both of these proteins are alginate specific | + | |
| - | + | ||
| + | ---- | ||
| + | ==Links to available structures== | ||
| + | Alginate binding periplasmic structures with binding to different alginate polymers | ||
| - | + | [http://www.rcsb.org/structure/1Y3N] :1Y3N | |
| - | [[ | + | [http://www.rcsb.org/structure/1Y3P] :1Y3P |
| - | By running through Consurf ([http://consurf.tau.ac.il/2016/]), the Phylogenetic tree of 1Y3N can be obtained. Below a link to this tree is provided. | + | |
| + | [http://www.rcsb.org/structure/1Y3Q] :1Y3Q | ||
| + | |||
| + | [http://www.rcsb.org/structure/1KWH] :1KWH | ||
| + | |||
| + | [http://www.rcsb.org/structure/1J1N] :1J1N | ||
| + | |||
| + | [http://www.rcsb.org/structure/3A09] :3A09 | ||
| + | |||
| + | [http://www.rcsb.org/structure/3VLU] :3VLU | ||
| + | |||
| + | [http://www.rcsb.org/structure/3VLV] :3VLV | ||
| + | |||
| + | [http://www.rcsb.org/structure/3VLW]: 3VLW | ||
| + | |||
| + | [http://www.rcsb.org/structure/5H6U] :5H6U | ||
| + | |||
| + | [http://www.rcsb.org/structure/5H71] | ||
| + | |||
| + | |||
| + | == Link to evolutionary related Structures== | ||
| + | |||
| + | By running the protein through Consurf ([http://consurf.tau.ac.il/2016/]), the Phylogenetic tree of 1Y3N can be obtained. Below a link to this tree is provided. | ||
[http://consurf.tau.ac.il//wasabi/?url=http://consurf.tau.ac.il/results/1525350845/query_msa_fasta_and_Tree.xml] | [http://consurf.tau.ac.il//wasabi/?url=http://consurf.tau.ac.il/results/1525350845/query_msa_fasta_and_Tree.xml] | ||
| + | The <scene name='55/559112/1y3q_evolutionary/1'>evolutionary conservation 3D structure of 1Y3Q (AlgQ1)</scene>is shown here. | ||
| + | |||
| + | |||
| + | ---- | ||
== References: == | == References: == | ||
<references /> | <references /> | ||
</StructureSection> | </StructureSection> | ||
Current revision
Alginate Binding Periplasmin Proteins of Sphingomonas sp. A1 (AlgQ1, AlgQ2)
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Julie Langlois, Atena Farhangian, Rebecca Holstein, Elizabeth A. Dunlap, Katherine Reynolds, Elizeu Santos, Noam Gonen, Anna Lohning, Idan Ben-Nachum, Brian Ochoa, Shai Biran, Gauri Misra, Shira Weingarten-Gabbay, Keni Vidilaseris, Jamie Costa, Abhinav Mittal, Urs Leisinger, Madison Walberry, Edmond R Atalla, Brett M. Thumm, Brooke Fenn, Joel L. Sussman, Mati Cohen, Vesta Nwankwo, Dotan Shaniv, Gulalai Shah