CRISPR-Cas
From Proteopedia
(Difference between revisions)
| (10 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='4qyz' size=' | + | <StructureSection load='4qyz' size='350' side='right' scene='74/742625/Cv/4' caption='E. coli CRISPR complex with RNA and DNA (PDB code [[4qyz]])'> |
'''PART I''' | '''PART I''' | ||
=Background= | =Background= | ||
| Line 8: | Line 8: | ||
*Gene networks can be efficiently probed and modified for biotechnology applications.<ref name="Did">PMID:27344519</ref> | *Gene networks can be efficiently probed and modified for biotechnology applications.<ref name="Did">PMID:27344519</ref> | ||
| - | CRISPR-Cas9 has recently emerged as a promising system for multiplexed genome editing as well as epigenome and transcriptome perturbation. Due to its specificity, ease of use and highly modular programmable nature, it has been widely adopted for a variety of applications such as genome editing, transcriptional inhibition and activation, genetic screening, DNA localization imaging, and many more. In this review, we will discuss non-editing applications of CRISPR-Cas9 for transcriptome perturbation, metabolic engineering, and synthetic biology.<ref name="Did">PMID:27344519</ref> | + | '''CRISPR-Cas9''' has recently emerged as a promising system for multiplexed genome editing as well as epigenome and transcriptome perturbation. Due to its specificity, ease of use and highly modular programmable nature, it has been widely adopted for a variety of applications such as genome editing, transcriptional inhibition and activation, genetic screening, DNA localization imaging, and many more. In this review, we will discuss non-editing applications of CRISPR-Cas9 for transcriptome perturbation, metabolic engineering, and synthetic biology.<ref name="Did">PMID:27344519</ref> |
Since the early days of genetic engineering there has been a need for control of gene expression. Naturally occurring transcription factors (TFs) have traditionally been used to achieve this goal (reviewed in <ref name="Prin1">PMID:24781324</ref>). However, their limited DNA binding sequence space required installing specific sequences within the transcription regulatory elements of the target genes. This can be technically difficult and may have unintended consequences on gene expression. Zinc fingers (ZFs) and transcription activator-like effectors (TALEs) were developed to overcome the fixed binding sequence requirements of native TFs. However, both ZFs and TALEs have significant limitations. ZFs have complicated design criteria and large highly repetitive TALE genes are difficult to synthesize and clone (reviewed in <ref name="Prin2">PMID:23718948</ref><ref name="Prin3">PMID:24584096</ref>). These challenges have recently been overcome using CRISPR-Cas9 based TFs. The biochemical properties of CRISPR-Cas9 based TFs that enable such flexibility and describe their applications to synthetic gene circuit design and multi-plexed perturbation of native gene networks.<ref name="Did">PMID:27344519</ref> | Since the early days of genetic engineering there has been a need for control of gene expression. Naturally occurring transcription factors (TFs) have traditionally been used to achieve this goal (reviewed in <ref name="Prin1">PMID:24781324</ref>). However, their limited DNA binding sequence space required installing specific sequences within the transcription regulatory elements of the target genes. This can be technically difficult and may have unintended consequences on gene expression. Zinc fingers (ZFs) and transcription activator-like effectors (TALEs) were developed to overcome the fixed binding sequence requirements of native TFs. However, both ZFs and TALEs have significant limitations. ZFs have complicated design criteria and large highly repetitive TALE genes are difficult to synthesize and clone (reviewed in <ref name="Prin2">PMID:23718948</ref><ref name="Prin3">PMID:24584096</ref>). These challenges have recently been overcome using CRISPR-Cas9 based TFs. The biochemical properties of CRISPR-Cas9 based TFs that enable such flexibility and describe their applications to synthetic gene circuit design and multi-plexed perturbation of native gene networks.<ref name="Did">PMID:27344519</ref> | ||
| Line 36: | Line 36: | ||
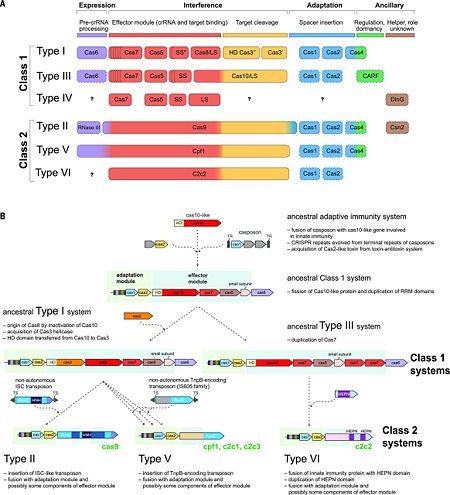
==CRISPR-Cas diversity, classification, and evolution== | ==CRISPR-Cas diversity, classification, and evolution== | ||
| - | The rapid evolution of highly diverse | + | '''Classification according to the Wikipedia page CRISPR [https://en.wikipedia.org/wiki/CRISPR] with additions''' |
| + | |||
| + | '''CRISPR Class 1 uses a complex of multiple Cas proteins''' | ||
| + | |||
| + | CRISPR type I (Cas3) | ||
| + | |||
| + | CRISPR type I-A (Cascade) - see [[CRISPR subtype I-A]] | ||
| + | |||
| + | CRISPR type I-B (Cascade) - see [[CRISPR subtype I-B]] | ||
| + | |||
| + | CRISPR type I-C (Cascade) - see [[CRISPR subtype I-C]] | ||
| + | |||
| + | CRISPR type I-D (Cas10d) | ||
| + | |||
| + | CRISPR type I-E (Cascade) - see [[CRISPR type I-E (Cascade)|CRISPR subtype I-E]] | ||
| + | |||
| + | CRISPR type I-F (Csy1, Csy2, Csy3) - see [[CRISPR subtype I-F]] | ||
| + | |||
| + | CRISPR type I-U (GSU0054) | ||
| + | |||
| + | CRISPR type III (Cas10) | ||
| + | |||
| + | CRISPR type III-A (Csm complex) - see [[CRISPR subtype III-A (Csm complex)]] | ||
| + | |||
| + | CRISPR type III-B (Cmr complex) | ||
| + | |||
| + | CRISPR type III-C (Cas10 or Csx11) | ||
| + | |||
| + | CRISPR type III-D (Csx10) | ||
| + | |||
| + | CRISPR type Orphan | ||
| + | |||
| + | CRISPR type IV (Csf1) | ||
| + | |||
| + | CRISPR type IV-A | ||
| + | |||
| + | CRISPR type IV-B | ||
| + | |||
| + | '''CRISPR Class 2 uses a single large Cas protein''' | ||
| + | |||
| + | CRISPR type II-A - see [[CRISPR-Cas9]] | ||
| + | |||
| + | CRISPR type II-B (Cas4) | ||
| + | |||
| + | CRISPR type II-C | ||
| + | |||
| + | CRISPR type V (Cpf1, C2c1, C2c3) - see [[CRISPR type V]] | ||
| + | |||
| + | CRISPR type VI (Cas13a (previously known as C2c2), Cas13b, Cas13c, Cas13d) - see [[CRISPR type VI]] | ||
| + | |||
| + | The rapid evolution of highly diverse CRISPR-Cas systems is thought to be driven by the continuous arms race with the invading MGEs. The latest classification scheme for CRISPR-Cas systems, which takes into account the repertoire of ''cas'' genes and the sequence similarity between Cas proteins and the locus architecture, includes two classes that are currently subdivided into six types and 19 subtypes <ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Rev430">doi:10.1038/nrmicro3569</ref><ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref>. The key feature of the organization and evolution of the CRISPR-Cas loci is their pronounced modularity. The module responsible for the adaptation step is largely uniform among the diverse CRISPR-Cas systems and consists of the ''cas1'' and ''cas2'' genes, both of which are essential for the acquisition of spacers. In many CRISPR-Cas variants, the adaptation module also includes the ''cas4'' gene. By contrast, the CRISPR-Cas effector module, which is involved in the maturation of the crRNAs as well as in target recognition and cleavage, shows a far greater versatility (Fig. 2A) <ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Rev430">doi:10.1038/nrmicro3569</ref>. | ||
[[Image:F2r.jpg|left|450px|thumb|Figure. 2. CRISPR diversity and evolution. (A) Modular organization of the CRISPR-Cas systems. LS, large subunit; SS, small subunit. A putative small subunit that might be fused to the large subunit in several type I subtypes is indicated by an asterisk. Cas3 is shown as fusion of two distinct genes encoding the helicase Cas3′ and the nuclease HD Cas3′′; in some type I systems, these domains are encoded by separate genes. Functionally dispensable components are indicated by dashed outlines. Cas6 is shown with a thin solid outline for type I because it is dispensable in some systems, and by a dashed line for type III because most systems lack this gene and use the Cas6 provided in trans by other CRISPR-Cas loci. The two colors for Cas4 and C2c2 and three colors for Cas9 and Cpf1 reflect the contributions of these proteins to different stages of the CRISPR-Cas response (see text). The question marks indicate currently unknown components. From <ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Rev430">doi:10.1038/nrmicro3569</ref> (B) Evolutionary scenario for the CRISPR-Cas systems. TR, terminal repeats; TS, terminal sequences; HD, HD-family endonuclease; HNH, HNH-family endonuclease; RuvC, RuvC-family endonuclease; HEPN, putative endoribonuclease of HEPN superfamily. Genes and portions of genes shown in gray denote sequences that are thought to have been encoded in the respective mobile elements but were eliminated in the course of evolution of CRISPR-Cas systems. From <ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref>]] | [[Image:F2r.jpg|left|450px|thumb|Figure. 2. CRISPR diversity and evolution. (A) Modular organization of the CRISPR-Cas systems. LS, large subunit; SS, small subunit. A putative small subunit that might be fused to the large subunit in several type I subtypes is indicated by an asterisk. Cas3 is shown as fusion of two distinct genes encoding the helicase Cas3′ and the nuclease HD Cas3′′; in some type I systems, these domains are encoded by separate genes. Functionally dispensable components are indicated by dashed outlines. Cas6 is shown with a thin solid outline for type I because it is dispensable in some systems, and by a dashed line for type III because most systems lack this gene and use the Cas6 provided in trans by other CRISPR-Cas loci. The two colors for Cas4 and C2c2 and three colors for Cas9 and Cpf1 reflect the contributions of these proteins to different stages of the CRISPR-Cas response (see text). The question marks indicate currently unknown components. From <ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Rev430">doi:10.1038/nrmicro3569</ref> (B) Evolutionary scenario for the CRISPR-Cas systems. TR, terminal repeats; TS, terminal sequences; HD, HD-family endonuclease; HNH, HNH-family endonuclease; RuvC, RuvC-family endonuclease; HEPN, putative endoribonuclease of HEPN superfamily. Genes and portions of genes shown in gray denote sequences that are thought to have been encoded in the respective mobile elements but were eliminated in the course of evolution of CRISPR-Cas systems. From <ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref>]] | ||
| Line 65: | Line 115: | ||
The prototype type V effector Cpf1 (subtype V-A) contains only one nuclease domain (RuvC-like) that is identifiable by sequence analysis. However, analysis of the recently solved structure of <scene name='74/746096/Cv5/1'>Cpf1 complexed with the crRNA and target DNA</scene> has revealed a second nuclease domain, the fold of which is unrelated to HNH or any other known nucleases. In analogy to the HNH domain in Cas9, the <scene name='74/746096/Cv5/2'>novel nuclease domain (labeled Nuc) in Cpf1 is inserted into the RuvC domain</scene>, and it is responsible for cleavage of the target strand.<ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Cpf1">PMID:24884953</ref> | The prototype type V effector Cpf1 (subtype V-A) contains only one nuclease domain (RuvC-like) that is identifiable by sequence analysis. However, analysis of the recently solved structure of <scene name='74/746096/Cv5/1'>Cpf1 complexed with the crRNA and target DNA</scene> has revealed a second nuclease domain, the fold of which is unrelated to HNH or any other known nucleases. In analogy to the HNH domain in Cas9, the <scene name='74/746096/Cv5/2'>novel nuclease domain (labeled Nuc) in Cpf1 is inserted into the RuvC domain</scene>, and it is responsible for cleavage of the target strand.<ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Cpf1">PMID:24884953</ref> | ||
| - | Screening of microbial genomes and metagenomes for undiscovered class 2 systems has resulted in the identification of three novel CRISPR-Cas variants. These include subtypes V-B and V-C, which resemble Cpf1 in that their predicted effector proteins contain a single, RuvC-like | + | Screening of microbial genomes and metagenomes for undiscovered class 2 systems has resulted in the identification of three novel CRISPR-Cas variants. These include subtypes V-B and V-C, which resemble Cpf1 in that their predicted effector proteins contain a single, RuvC-like nuclease domain. Cleavage of target DNA by the type V-B effector, denoted C2c1, has been experimentally demonstrated. Type VI is unique in that its effector protein contains two conserved HEPN domains that possess ribonuclease (RNase) activity (Fig. 2A).<ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref> |
| - | nuclease domain. Cleavage of target DNA by the type V-B effector, denoted C2c1, has been experimentally demonstrated. Type VI is unique in that its effector protein contains two conserved HEPN domains that possess ribonuclease (RNase) activity (Fig. 2A).<ref name="Rev4">doi:10.1126/science.aad5147</ref><ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref> | + | |
Recent comparative genomic analyses of variant CRISPR-Cas systems (Fig. 2B) <ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref> have revealed a strong modular evolution with multiple combinations of adaptation modules and effector modules, as well as a pivotal contribution of mobile genetic elements to the origin and diversification of the CRISPR-Cas systems. The ancestral prokaryotic adaptive immune system could have emerged via the insertion of a casposon (a recently discovered distinct class of self-synthesizing transposons that appear to encode a Cas1 homolog) next to an innate immunity locus (probably consisting of genes encoding a Cas10 nuclease and possibly one or more RNA binding proteins). Apart from providing the Cas1 nuclease/integrase that is required for recombination during spacer acquisition, the casposon may also have contributed the prototype CRISPR repeat unit that could have evolved from one of the inverted terminal repeats of the casposon. An additional toxin-antitoxin module that inserted either in the ancestral casposon or in the evolving adaptive immunity locus probably provided the ''cas2'' gene, thus completing the adaptation module. The Cas10 nuclease and one or more additional proteins with an RRM fold (the ultimate origin of which could be a polymerase or cyclase that gave rise to Cas10) of the hybrid locus could have subsequently evolved to become the ancestral CRISPR-Cas effector module <ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref><ref name="Rev4">doi:10.1126/science.aad5147</ref>. | Recent comparative genomic analyses of variant CRISPR-Cas systems (Fig. 2B) <ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref> have revealed a strong modular evolution with multiple combinations of adaptation modules and effector modules, as well as a pivotal contribution of mobile genetic elements to the origin and diversification of the CRISPR-Cas systems. The ancestral prokaryotic adaptive immune system could have emerged via the insertion of a casposon (a recently discovered distinct class of self-synthesizing transposons that appear to encode a Cas1 homolog) next to an innate immunity locus (probably consisting of genes encoding a Cas10 nuclease and possibly one or more RNA binding proteins). Apart from providing the Cas1 nuclease/integrase that is required for recombination during spacer acquisition, the casposon may also have contributed the prototype CRISPR repeat unit that could have evolved from one of the inverted terminal repeats of the casposon. An additional toxin-antitoxin module that inserted either in the ancestral casposon or in the evolving adaptive immunity locus probably provided the ''cas2'' gene, thus completing the adaptation module. The Cas10 nuclease and one or more additional proteins with an RRM fold (the ultimate origin of which could be a polymerase or cyclase that gave rise to Cas10) of the hybrid locus could have subsequently evolved to become the ancestral CRISPR-Cas effector module <ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref><ref name="Rev4">doi:10.1126/science.aad5147</ref>. | ||
The widespread occurrence of class 1 systems in archaea and bacteria, together with the proliferation of the ancient RRM domain in class 1 effector proteins, strongly suggests that the ancestral CRISPR-Cas belonged to class 1. Most likely, the multiple class 2 variants then evolved via several independent replacements of the class 1 effector locus with nuclease genes that were derived from distinct MGEs (Fig. 2B). In particular, type V effector variants (Cpf1) seem to have evolved from different families of the TnpB transposase genes that are widespread in transposons <ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref>, whereas the type II effector (Cas9) may have evolved from IscB, a protein with two nuclease domains that belongs to a recently identified distinct transposon family. Notably, class 2 CRISPR-Cas systems, in their entirety, appear to have been derived from different MGEs: Cas1 from a casposon, Cas2 from a toxin-antitoxin module, and the different effector proteins (such as Cas9 and Cpf1) from respective transposable elements <ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref><ref name="Rev4">doi:10.1126/science.aad5147</ref>. | The widespread occurrence of class 1 systems in archaea and bacteria, together with the proliferation of the ancient RRM domain in class 1 effector proteins, strongly suggests that the ancestral CRISPR-Cas belonged to class 1. Most likely, the multiple class 2 variants then evolved via several independent replacements of the class 1 effector locus with nuclease genes that were derived from distinct MGEs (Fig. 2B). In particular, type V effector variants (Cpf1) seem to have evolved from different families of the TnpB transposase genes that are widespread in transposons <ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref>, whereas the type II effector (Cas9) may have evolved from IscB, a protein with two nuclease domains that belongs to a recently identified distinct transposon family. Notably, class 2 CRISPR-Cas systems, in their entirety, appear to have been derived from different MGEs: Cas1 from a casposon, Cas2 from a toxin-antitoxin module, and the different effector proteins (such as Cas9 and Cpf1) from respective transposable elements <ref name="Rev431">doi:10.1016/j.molcel.2015.10.008</ref><ref name="Rev4">doi:10.1126/science.aad5147</ref>. | ||
| - | |||
| - | ==CRISPR adaptation== | ||
| - | |||
| - | The spacers of a CRISPR array represent a chronological archive of previous invader encounters. The captured spacer sequences are integrated into the CRISPR loci after exposure to MGEs, at the leader end of the array that contains the start site of CRISPR transcription. Analysis of invader target sequences (also called protospacers) has revealed a short motif directly adjacent to the target sequence, called the protospacer adjacent motif (PAM). This PAM motif allows self/nonself discrimination by the host in two ways: (i) because its presence in alien targets is required for nonself interference, and (ii) because its absence in the host’s CRISPR array avoids self-targeting. In class 1–type I and class 2–type II systems, the PAM is not only involved in interference, but also plays a role in spacer selection during the adaptation stage, implying the acquisition of functional spacers only. The PAM is a short [2 to 7 nucleotides (nt)], partially redundant sequence that in itself cannot preclude incorporation of spacers from the host DNA because of the low information content of the motif. The short PAM appears to be the result of an evolutionary trade-off between efficient incorporation of spacers from nonself DNA and preventing an autoimmune reaction.<ref name="Rev4">doi:10.1126/science.aad5147</ref> | ||
| - | |||
| - | ''Examples of PAM:'' | ||
| - | *<scene name='74/746096/Cv6/3'>PAM-complementary dual-forked DNA, which is a 23-mer palindromic duplex</scene> from ''Escherichia coli'' ([[5dqz]]). | ||
| - | *<scene name='74/742625/Cv2/10'>PAM in crystal structure of Acidaminococcus sp. Cpf1 in complex with crRNA and target DNA</scene> ([[5b43]]). | ||
| - | *<scene name='74/742625/Cv2/9'>PAM in crRNA-dsDNA hybrid from E. coli</scene> ([[5h9f]]). | ||
| - | *<scene name='74/742625/Cv/42'>PAM in Cas9-sgRNA-target DNA complex from Streptococcus pyogenes</scene> ([[5fw2]]). | ||
| - | *<scene name='74/742625/Cv2/13'>PAM in Cas9-sgRNA-target DNA complex from Francisella tularensis</scene> ([[5b2p]]). | ||
| - | *<scene name='74/742625/Cv3/2'>PAM in Cas9-sgRNA-target DNA complex from Staphylococcus aureus</scene> ([[4axw]]). | ||
| - | *<scene name='74/742625/Cv3/10'>Few nucleotide long conserved motif recognized directly by Cas9 protein (protospacer adjacent motif, PAM)</scene>. | ||
| - | |||
| - | Although host chromosomal fragments can be incorporated as new CRISPR spacers, detection of such events obviously implies that this did not result in a lethal phenotype, either due to a modified PAM and/or to an inactivated CRISPR-Cas effector module <ref name="Rev451">doi:10.1016/ | ||
| - | j.tig.2010.05.008</ref>. Indeed, in the absence of the effector module, elevated frequencies of self-spacer acquisition occur in ''Escherichia coli'' <ref name="Rev452">doi:10.1093/nar/gks216</ref>. Similarly, ''Streptococcus thermophilus'' with a catalytically inactive Cas9 results in a major increase of spacers derived from the host genome <ref name="Rev453">doi:10.1101/gad.257550.114</ref>. In addition, there is a strong preference for the integration of plasmid over chromosomal spacer sequences <ref name="Rev452">doi:10.1093/nar/gks216</ref>, with plasmid sequences incorporated more frequently than host DNA by two to three orders of magnitude <ref name="Rev456">doi:10.1038/nature14302</ref>. Spacer acquisition in ''E. coli'' requires active replication of the protospacercontaining DNA <ref name="Rev456">doi:10.1038/nature14302</ref>. Thus, small, fast-replicating plasmid genomes are a much better source of spacers than the large host DNA, and such findings are consistent with acquisition of spacers from an infecting virus genome in the archaeon ''Sulfolobus islandicus'' requiring its active replication <ref name="Rev457">doi:10.1111/mmi.12503</ref>. In ''E. coli'', the CRISPR-Cas system derives the spacers primarily from products of RecBCD-catalyzed DNA degradation that are formed during the repair of double-stranded breaks associated with stalled replication forks <ref name="Rev458">doi:10.1016/j.cell.2007.11.004</ref>. Other possible sources of substrates for CRISPR adaptation include DNA fragments generated | ||
| - | either by other defense systems, such as restriction-modification systems <ref name="Rev459">doi:10.1038/ncomms3087</ref>, or by the CRISPR-Cas system itself <ref name="Rev449">doi:10.1371/journal.pone.0035888</ref>.<ref name="Rev4">doi:10.1126/science.aad5147</ref> | ||
| - | |||
| - | <scene name='74/746096/Cv6/4'>Cas1 and Cas2</scene> play crucial roles in spacer acquisition in all CRISPR-Cas systems <ref name="Rev452">doi:10.1093/nar/gks216</ref>. In addition, these proteins can function in trans, provided that the repeats involved are sufficiently similar in size and structure. Accordingly, ''cas1'' and ''cas2'' genes are missing in many active CRISPR-Cas loci—in particular, of type III as well as types IV and VI <ref name="Rev430">doi:10.1038/nrmicro3569</ref>. Overexpression of Cas1 and Cas2 from the ''E. coli'' type I-E system has been shown to be sufficient for the extension of the CRISPR array <ref name="Rev452">doi:10.1093/nar/gks216</ref>. Mutations in the active site of Cas1 abolish spacer integration in ''E. coli'' <ref name="Rev452">doi:10.1093/nar/gks216</ref>, whereas the nuclease activity of Cas2 is dispensable <ref name="Rev455">doi:10.1038/nsmb.2820</ref>. In ''E. coli'', a <scene name='74/746096/Cv6/5'>central Cas2 dimer and two flanking Cas1 dimers form a complex that binds and processes PAM containing DNA fragments</scene> ([[5dqz]]; Fig. 3A <ref name="Rev455">doi:10.1038/nsmb.2820</ref>, <ref name="Rev460">doi:10.1016/j.cell.2015.10.008</ref>), after which the newly generated spacers can be integrated into a CRISPR array via a recombination mechanism akin to that of retroviral integrases and transposases <ref name="Rev461">doi:10.1038/nature14237</ref> (Fig. 3B). <scene name='74/746096/Cv6/7'>Cas1 cleaves the phosphodiester bond between nucleotides 28 and 29</scene>, resulting in a DNA cleavage product. <scene name='74/746096/Cv6/8'>Glu141, His208, and Asp221 are the catalytic residues of Cas1</scene>. The DNA cleavage site is labeled in red. | ||
| - | |||
| - | [[Image:F4large.jpg|left|450px|thumb|Fig. 3 Spacer acquisition. (A) Crystal structure of the complex of Cas1-Cas2 bound to the dual-forked DNA (PDB accession [[5dqz]]). The target DNA is shown in dark blue; the Cas1 and Cas2 dimers of the complex are indicated in blue and yellow, respectively. (B) Model explaining the capture of new DNA sequences from invading nucleic acid and the subsequent DNA integration into the host CRISPR array. The numbers on the left correspond to the order of events as described in the text. The dashed lines indicate nucleotides; the nucleotides C and N on the two sides of the protospacer are shown in red and green to clarify the orientation. From <ref name="Rev4">doi:10.1126/science.aad5147</ref>]] | ||
| - | {{Clear}} | ||
| - | |||
| - | In several type III CRISPR-Cas systems, Cas1 is fused to reverse transcriptase (20), and it was recently shown that these systems are capable of acquisition of RNA spacers by direct incorporation of an RNA segment into the CRISPR array followed by reverse transcription and replacement of the RNA strand by DNA <ref name="Rev462">doi:10.1126/science.aad4234</ref>. Although the biological function of this process remains to be elucidated, these findings demonstrate remarkable versatility of adaptation pathways. Spacer acquisition (adaptation) in type I systems proceeds along two distinct paths: (i) naïve acquisition, which occurs during an initial infection, and (ii) primed acquisition, when the CRISPR contains a previously integrated spacer that is complementary to the invading DNA <ref name="Rev463">doi:10.1016/j.virol.2012.10.003</ref>. According to the proposed model, naïve spacer adaptation involves five steps (Fig. 3B): | ||
| - | 1) Fragmentation of (mainly) invasive nucleic acids by non-Cas systems [e.g., by RecBCD after stalling a replication fork, or by restriction enzymes (restriction-modification systems) <ref name="Rev456">doi:10.1038/nature14302</ref><ref name="Rev459">doi:10.1038/ncomms3087</ref>] or by CRISPR-associated nucleases <ref name="Rev449">doi:10.1371/journal.pone.0035888</ref>. Although this step may be non-essential, it probably enhances the efficiency of the overall process and its specificity toward invading DNA. | ||
| - | 2) Selection of DNA fragments for (proto) spacers by scanning for potential PAMs (after partial target unwinding) by one of the four Cas1 subunits of the Cas1-Cas2 complex <ref name="Rev464">doi:10.1093/nar/gku510</ref>. | ||
| - | 3) Measuring of the selected protospacer generating fragments of the correct size with 3′ hydroxyl groups by Cas1 nuclease. | ||
| - | 4) Nicking of both strands of the leaderproximal repeat of the CRISPR array at the 5′ ends through a direct nucleophilic attack by the generated 3′ OH groups, resulting in covalent links of each of the strands of the newly selected spacer to the single-stranded repeat ends. | ||
| - | 5) Second-strand synthesis and ligation of the repeat flanks by a non-Cas repair system <ref name="Rev446">doi:10.1038/nrmicro.2015.14</ref><ref name="Rev461">doi:10.1038/nature14237</ref>.<ref name="Rev4">doi:10.1126/science.aad5147</ref> | ||
SEE [[CRISPR-Cas Part II]] | SEE [[CRISPR-Cas Part II]] | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Didovyk A, Borek B, Tsimring L, Hasty J. Transcriptional regulation with CRISPR-Cas9: principles, advances, and applications. Curr Opin Biotechnol. 2016 Aug;40:177-84. doi: 10.1016/j.copbio.2016.06.003. Epub, 2016 Jun 23. PMID:27344519 doi:http://dx.doi.org/10.1016/j.copbio.2016.06.003
- ↑ Brophy JA, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014 May;11(5):508-20. doi: 10.1038/nmeth.2926. PMID:24781324 doi:http://dx.doi.org/10.1038/nmeth.2926
- ↑ Straubeta A, Lahaye T. Zinc fingers, TAL effectors, or Cas9-based DNA binding proteins: what's best for targeting desired genome loci? Mol Plant. 2013 Sep;6(5):1384-7. doi: 10.1093/mp/sst075. Epub 2013 May 29. PMID:23718948 doi:http://dx.doi.org/10.1093/mp/sst075
- ↑ Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014 Apr;32(4):347-55. doi: 10.1038/nbt.2842. Epub 2014 Mar 2. PMID:24584096 doi:http://dx.doi.org/10.1038/nbt.2842
- ↑ 5.0 5.1 5.2 5.3 5.4 Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015 Jan;40(1):58-66. doi: 10.1016/j.tibs.2014.10.007. Epub, 2014 Nov 18. PMID:25468820 doi:http://dx.doi.org/10.1016/j.tibs.2014.10.007
- ↑ 6.0 6.1 Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007 Mar 23;315(5819):1709-12. PMID:17379808 doi:http://dx.doi.org/10.1126/science.1138140
- ↑ 7.00 7.01 7.02 7.03 7.04 7.05 7.06 7.07 7.08 7.09 7.10 7.11 7.12 7.13 7.14 Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016 Aug 5;353(6299):aad5147. doi: 10.1126/science.aad5147. PMID:27493190 doi:http://dx.doi.org/10.1126/science.aad5147
- ↑ Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8(4):R61. PMID:17442114 doi:http://dx.doi.org/10.1186/gb-2007-8-4-r61
- ↑ 9.0 9.1 9.2 Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008 Aug 15;321(5891):960-4. doi: 10.1126/science.1159689. PMID:18703739 doi:http://dx.doi.org/10.1126/science.1159689
- ↑ Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010 Nov 4;468(7320):67-71. doi: 10.1038/nature09523. PMID:21048762 doi:http://dx.doi.org/10.1038/nature09523
- ↑ 11.0 11.1 11.2 11.3 11.4 Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015 Nov;13(11):722-36. doi: 10.1038/nrmicro3569. Epub 2015, Sep 28. PMID:26411297 doi:http://dx.doi.org/10.1038/nrmicro3569
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 12.6 12.7 12.8 Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015 Nov 5;60(3):385-97. doi: 10.1016/j.molcel.2015.10.008. Epub 2015, Oct 22. PMID:26593719 doi:http://dx.doi.org/10.1016/j.molcel.2015.10.008
- ↑ Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015 Jun 26;348(6242):1477-81. doi: 10.1126/science.aab1452. PMID:26113724 doi:http://dx.doi.org/10.1126/science.aab1452
- ↑ 14.0 14.1 14.2 Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816-21. doi: 10.1126/science.1225829. Epub 2012, Jun 28. PMID:22745249 doi:http://dx.doi.org/10.1126/science.1225829
- ↑ Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):E2579-86. Epub 2012 Sep 4. PMID:22949671 doi:http://dx.doi.org/10.1073/pnas.1208507109
- ↑ Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013 Feb 28;152(5):1173-83. doi: 10.1016/j.cell.2013.02.022. PMID:23452860 doi:http://dx.doi.org/10.1016/j.cell.2013.02.022
- ↑ Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013 Aug;41(15):7429-37. doi: 10.1093/nar/gkt520. Epub 2013, Jun 12. PMID:23761437 doi:http://dx.doi.org/10.1093/nar/gkt520
- ↑ Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014 Jul;32(7):677-83. doi: 10.1038/nbt.2916. Epub 2014 May 18. PMID:24837660 doi:http://dx.doi.org/10.1038/nbt.2916
- ↑ Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014 May 19;12:36. doi: 10.1186/1741-7007-12-36. PMID:24884953 doi:http://dx.doi.org/10.1186/1741-7007-12-36
Categories: Topic Page | Crispr | Crispr-associated | Endonuclease | Cas9 | Cas6
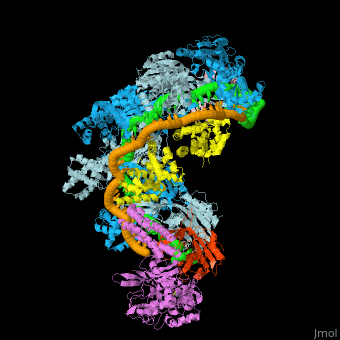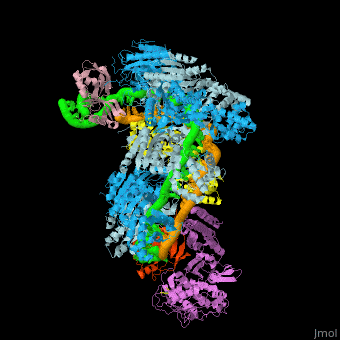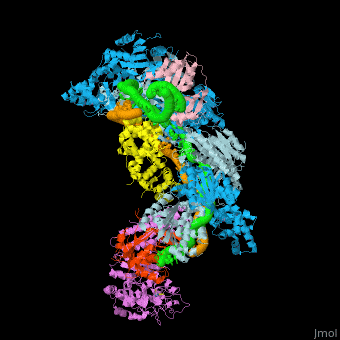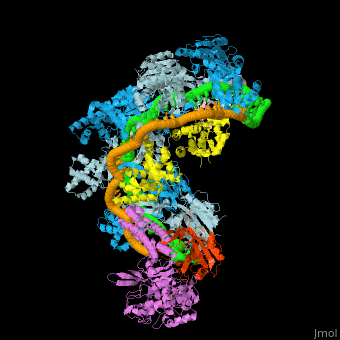
![Fig. 1 Overview of the CRISPR-Cas systems. (A) Architecture of class 1 (multiprotein effector complexes) and class 2 (single-protein effector complexes) CRISPR-Cas systems. (B) CRISPR-Cas adaptive immunity is mediated by CRISPR RNAs (crRNAs) and Cas proteins, which form multicomponent CRISPR ribonucleoprotein (crRNP) complexes. The first stage is adaptation, which occurs upon entry of an invading mobile genetic element (in this case, a viral genome). Cas1 (blue) and Cas2 (yellow) proteins select and process the invading DNA, and thereafter, a protospacer (orange) is integrated as a new spacer at the leader end of the CRISPR array [repeat sequences (gray) that separate similar-sized, invader-derived spacers (multiple colors)]. During the second stage, expression, the CRISPR locus is transcribed and the pre-crRNA is processed into mature crRNA guides by Cas (e.g., Cas6) or non-Cas proteins (e.g., RNase III). During the final interference stage, the Cas-crRNA complex scans invading DNA for a complementary nucleic acid target, after which the target is degraded by a Cas nuclease. From](/wiki/images/thumb/f/f2/F1r.jpg/450px-F1r.jpg)