Sandbox Reserved 1502
From Proteopedia
(Difference between revisions)
| (42 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | {{Sandbox_Reserved_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | + | <scene name='80/802676/Site_of_interaction/1'>Text To Be Displayed</scene>{{Sandbox_Reserved_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> |
=='''3lpt - HIV integrase'''== | =='''3lpt - HIV integrase'''== | ||
<StructureSection load='3lpt' size='340' side='right' caption='[[3lpt]], [[Resolution|resolution]] 2.00Å' scene=''> | <StructureSection load='3lpt' size='340' side='right' caption='[[3lpt]], [[Resolution|resolution]] 2.00Å' scene=''> | ||
| - | + | The 3lpt is an integrase of the HIV, the Human Immunodeficiency Virus. An integrase is an enzyme produced by a retrovirus to integrate its genetic material into the DNA of the infected cell. It is one of three enzymes of HIV, the others being the Reverse Transcriptase and the Protease. | |
== Structural highlights == | == Structural highlights == | ||
| + | |||
| + | |||
| + | <scene name='80/802676/Hiv_integrase_3lpt/8'>HIV Integrase 3D structure</scene> | ||
| + | |||
<table><tr><td colspan='2'> | <table><tr><td colspan='2'> | ||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=723:(6-CHLORO-2-OXO-4-PHENYL-1,2-DIHYDROQUINOLIN-3-YL)ACETIC+ACID'>723</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=P03:2-[3-[3-(2-HYDROXYETHOXY)PROPOXY]PROPOXY]ETHANOL'>P03</scene>, <scene name='pdbligand=PEG:DI(HYDROXYETHYL)ETHER'>PEG</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=723:(6-CHLORO-2-OXO-4-PHENYL-1,2-DIHYDROQUINOLIN-3-YL)ACETIC+ACID'>723</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=P03:2-[3-[3-(2-HYDROXYETHOXY)PROPOXY]PROPOXY]ETHANOL'>P03</scene>, <scene name='pdbligand=PEG:DI(HYDROXYETHYL)ETHER'>PEG</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | ||
| Line 12: | Line 16: | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3lpt FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3lpt OCA], [http://pdbe.org/3lpt PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=3lpt RCSB], [http://www.ebi.ac.uk/pdbsum/3lpt PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=3lpt ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3lpt FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3lpt OCA], [http://pdbe.org/3lpt PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=3lpt RCSB], [http://www.ebi.ac.uk/pdbsum/3lpt PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=3lpt ProSAT]</span></td></tr> | ||
</table> | </table> | ||
| + | |||
== Function == | == Function == | ||
| - | + | An integrase is an enzyme required for the integration of viral DNA into the host genome. The process of integration can be divided into two sequential reactions. In the first one, named 3'-processing, the enzyme removes di- or trinucleotides from viral DNA ends to expose 3′-hydroxyls attached to CA dinucleotides. The second step is the insertion of the processed 3′-viral DNA ends into the host chromosomal DNA by a trans-esterification reaction. This is called the strand transfer. | |
| + | The HIV integrase is the enzyme responsible for the integration of the HIV's viral DNA into the host cell. | ||
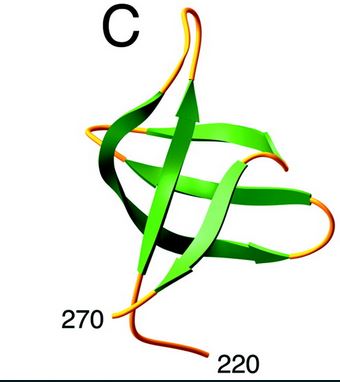
| - | == | + | == Structure == |
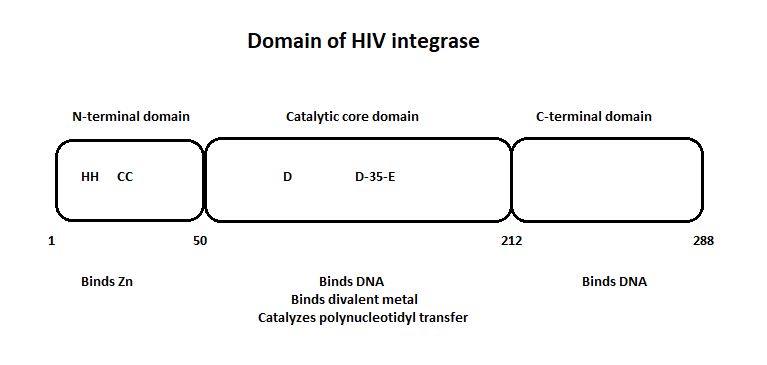
| - | + | HIV-1 integrase is composed of three domains: the N-terminal (residues 1-49), the core domain (residues 50-212) and the C-terminal domain (residues 213-288).[http://www.jbc.org/content/276/26/23213/F2.expansion.html] | |
| - | + | ||
| + | '''The three domain of the HIV integrase:''' | ||
| - | + | [[Image:Domain_HIV_Integrase.jpg]] [http://www.jbc.org/content/276/26/23213.full] | |
| - | HIV | + | '''HIV Integrase sequence:''' |
| - | + | [[Image:HIV integrase sequenc.jpg]][https://www.uniprot.org/uniprot/Q76353#sequences] | |
| + | Length:288, Mass (Da):32,199 | ||
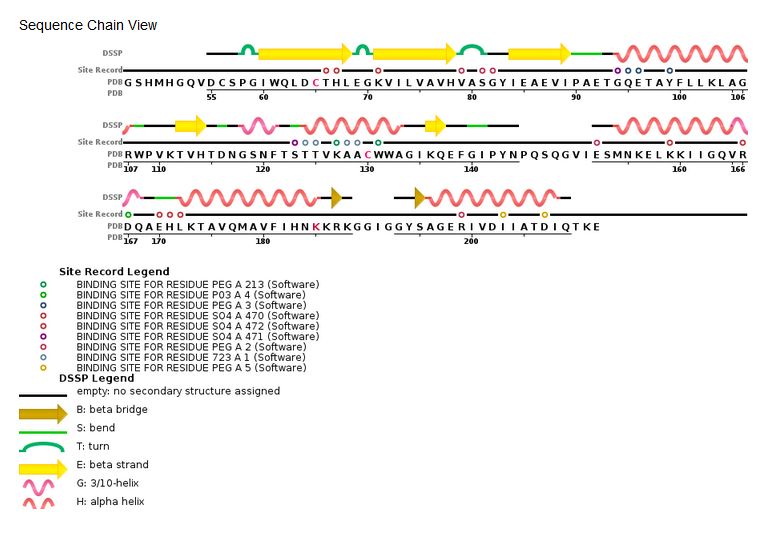
| - | ''' | + | '''Sequence chain view of the HIV Integrase:''' |
| - | [[Image: | + | [[Image:HIV Integrase sequence.jpg]] [http://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=3lpt] |
| - | + | ====Structure and role of the core domain==== | |
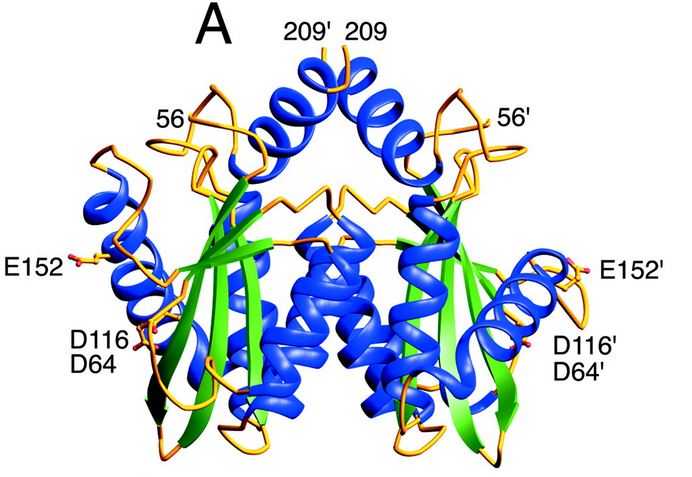
| - | + | The catalytic core domain contains three acidic residues, the D,D-35-E motif, comprising residues Asp64, Asp116, and Glu152. By analogy with models of catalysis by DNA polymerases, it has been proposed that coordination of divalent metal ion to these residues plays a key role in catalysis. The catalytic residues Asp64, Asp116, and Glu152 of HIV integrase are in close proximity, coordinate divalent metal ion, and define the active site. The residues comprising the active site region exhibit considerable flexibility. That suggests that binding of DNA substrate is required to impose the precise configuration of residues that are required for catalysis. | |
| - | [[Image: | + | [[Image:Core enzyme.jpg]] [http://www.jbc.org/content/276/26/23213.full] |
| - | = | + | <scene name='80/802676/Core_enzyme/2'>Here is the 3D structure of the core domain</scene> |
| - | The | + | |
| - | The | + | ====Structure and role of the N ter domain==== |
| + | |||
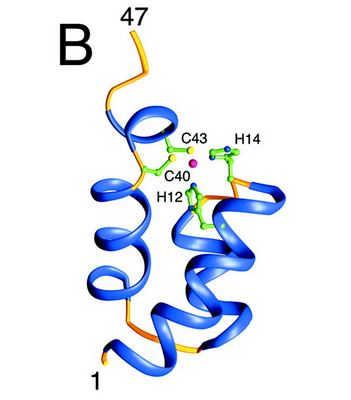
| + | The N-terminal domain of HIV-1 integrase contains a conserved pair of His and Cys residues, a motif similar to the zinc-coordinating residues of zinc fingers. This domain binds zinc but its structure is totally different from that of zinc fingers. It consists of a bundle of three α-helices. This domain is necessary for the oligomerization of the integrase. | ||
| + | |||
| + | [[Image:Nter-domain.jpg]] [http://www.jbc.org/content/276/26/23213.full] | ||
| + | |||
| + | <scene name='80/802676/N-ter_domain/2'>Here is the 3D structure of the N-ter domain of the HIV integrase</scene> | ||
| + | <table><tr><td colspan='2'></td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | ||
| + | </table> | ||
| + | |||
| + | ====Structure and role of the C ter domain==== | ||
| + | |||
| + | Here is the structure of the C-ter domain : | ||
| + | |||
| + | [[Image:C-ter domain.jpg]] [http://www.jbc.org/content/276/26/23213.full] | ||
| + | |||
| + | The C-terminal domain interacts in a non-specific way with DNA and would, therefore, play a stabilizing role of integrase-ADN interactions. | ||
| + | |||
| + | |||
| + | == Interaction between the HIV integrase and LEDGF/p75 == | ||
| + | |||
| + | LEDGF/p75, or Lens epithelium-derived growth factor, is a cellular cofactor of HIV integrase. It promotes the viral integration of the retrovirus by tethering the preintegration complex to the chromatin. Research on the LEDGF/p75-integrase interaction [https://www.nature.com/articles/nchembio.370] shows that it can be inhibited by using a molecule which will take the place of LEDGF/p75 on its binding site. Thus, the viral DNA will not be inserted into the genome of the host cell and the replication of HIV won't be possible. | ||
| + | |||
| + | ====Use of antiviral to inhibit the LEDGF/p75-integrase interaction ==== | ||
| + | |||
| + | An antiviral refers to a molecule disrupting the replication cycle of one or more viruses, thus allowing to slow down but rarely to stop a viral infection. Antiviral targeting specific regions of the integrase are used to prevent the replication of HIV. For example, the 2-(quinolin-3-yl)acetic acid interacts with <scene name='80/802676/Site_of_interaction/7'>the binding site of the LEDGF/p75 cofactor</scene>. More precisely, it binds to the residues E170, H171, and T174. This site is situated in the core domain of the enzyme. Once it is fixed into the core domain, the LEDGF/p75 will not be able to join and viral integration of the retrovirus will not be possible anymore. | ||
| + | The whole complex between the integrase and the cofactor is displayed on the Proteopedia page [[2b4j]]. | ||
== HIV == | == HIV == | ||
| - | HIV stands for human immunodeficiency virus. It is a retrovirus virus that damages the cells in the immune system and weakens the ability to fight everyday infections and disease. Copied into DNA, HIV is inserted into the genome of the infected cell thanks to integrases. To replicate and diffuse himself it attacks the body’s immune system, specifically the CD4 cells (T cells), which help fight off infections. As soon as HIV enters an individual, it accumulates in | + | HIV stands for human immunodeficiency virus. It is a retrovirus virus that damages the cells in the immune system and weakens the ability to fight everyday infections and disease. Copied into DNA, HIV is inserted into the genome of the infected cell thanks to integrases. To replicate and diffuse himself it attacks the body’s immune system, specifically the CD4 cells (T cells), which help fight off infections. As soon as HIV enters an individual, it accumulates in the cells and forms reservoirs of latent viruses in a few days or even hours. These reservoirs persist for life. |
| + | |||
| + | No cure currently exists, but HIV can be controlled by using antiretroviral therapy or ART. It is a combination of antiretroviral drugs to maximally suppress the HIV virus and stop the progression of HIV disease. In the mid-1990s, before the introduction of ART, people with HIV could progress to AIDS in just a few years. Today, someone diagnosed with HIV and treated before the disease is far advanced can live nearly as long as someone who does not have HIV.[https://www.hiv.gov/hiv-basics/overview/about-hiv-and-aids/what-are-hiv-and-aids] | ||
| + | |||
| + | == AIDS == | ||
| + | |||
| + | AIDS is the ultimate stage of infection with the human immunodeficiency virus. The word AIDS stands for Acquired Immunodeficiency Syndrome. People with AIDS get an increasing number of severe illnesses, called opportunistic infections, because of there damaged immune system. Are considered to have progressed to AIDS individual whose number of CD4 cells is lower than 200 cells per cubic millimeter of blood (200 cells/mm3). CD4 counts are between 500 and 1,600 cells/mm3 for someone with a healthy immune system. Developing one or more opportunistic illnesses, regardless of the CD4 count is also an indicator showing the progression to AIDS.[https://www.hiv.gov/hiv-basics/overview/about-hiv-and-aids/what-are-hiv-and-aids] | ||
| - | No cure currently exists, but HIV can be controlled by using antiretroviral therapy or ART. It is a combination of antiretroviral drugs to maximally suppress the HIV virus and stop the progression of HIV disease. Before the introduction of ART in the mid-1990s, people with HIV could progress to AIDS in just a few years. Today, someone diagnosed with HIV and treated before the disease is far advanced can live nearly as long as someone who does not have HIV.[https://www.hiv.gov/hiv-basics/overview/about-hiv-and-aids/what-are-hiv-and-aids] | ||
| - | == Disease: AIDS == | ||
| - | + | ==See also== | |
| + | http://proteopedia.org/wiki/index.php/2b4j :integrase ligated with the cofactor LEDGF/p75 | ||
| - | <scene name='80/802676/Hiv_integrase_3lpt/8'>HIV Integrase 3lpt</scene> | ||
== References == | == References == | ||
https://www.hiv.gov/hiv-basics/overview/about-hiv-and-aids/what-are-hiv-and-aids | https://www.hiv.gov/hiv-basics/overview/about-hiv-and-aids/what-are-hiv-and-aids | ||
https://www-nature-com.scd-rproxy.u-strasbg.fr/articles/nchembio.370 | https://www-nature-com.scd-rproxy.u-strasbg.fr/articles/nchembio.370 | ||
https://www.uniprot.org/uniprot/Q76353#sequences | https://www.uniprot.org/uniprot/Q76353#sequences | ||
| + | http://pfam.xfam.org/family/rve | ||
| + | http://pfam.xfam.org/protein/Q76353_9HIV1 | ||
| + | https://www.rcsb.org/3d-view/3LPT/1 | ||
| + | https://en.wikipedia.org/wiki/Diethylene_glycol | ||
| + | https://microbewiki.kenyon.edu/index.php/HIV-1_Pre-Integration_Complex | ||
| + | https://www.uniprot.org/uniprot/Q76353 | ||
| + | https://www.rcsb.org/structure/3lpt | ||
| + | http://proteopedia.org/wiki/index.php/Retroviral_Integrase | ||
| + | http://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=3lpt | ||
<references/> | <references/> | ||
Current revision
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
3lpt - HIV integrase
| |||||||||||