Bacterial thiolase
From Proteopedia
(Difference between revisions)
| Line 78: | Line 78: | ||
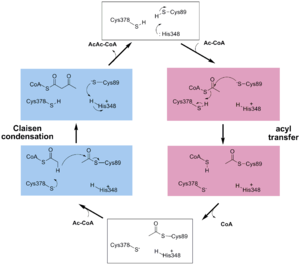
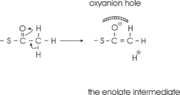
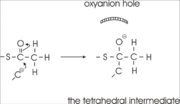
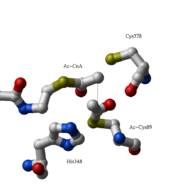
site geometry: a Asn316-Wat82 dyad, together with His348 make <scene name='User:Satyan_Sharma/Sandbox_1/Oxyanionhole_1/2'>oxyanion hole 1</scene>, stabilizing the CoA-thioester enolate intermediate. The hydrogen bond donors of oxyanion hole 1 are Wat82 and NE2 (His348). This enolate is formed from acetyl-CoA after proton | site geometry: a Asn316-Wat82 dyad, together with His348 make <scene name='User:Satyan_Sharma/Sandbox_1/Oxyanionhole_1/2'>oxyanion hole 1</scene>, stabilizing the CoA-thioester enolate intermediate. The hydrogen bond donors of oxyanion hole 1 are Wat82 and NE2 (His348). This enolate is formed from acetyl-CoA after proton | ||
abstraction by the catalytic base, Cys378 (Figure 3). | abstraction by the catalytic base, Cys378 (Figure 3). | ||
| - | <applet load='1dm3' size='300' frame='true' align='right' caption='The bacterial biosynthetic thiolase showing the acetyl CoA complex with sulfate: [[1DM3]], resolution 2.00 Å.'/> | ||
| Line 143: | Line 142: | ||
''Zoogloea ramigera'' is indeed important for the function of oxyanion | ''Zoogloea ramigera'' is indeed important for the function of oxyanion | ||
hole 1. | hole 1. | ||
| - | </StructureSection | + | </StructureSection> |
==Additional Resources== | ==Additional Resources== | ||
For additional information, see: [[Metabolic Disorders]] | For additional information, see: [[Metabolic Disorders]] | ||
Revision as of 09:47, 4 February 2019
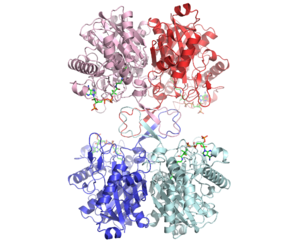
3D structure (1DM3) of the bacterial Zoogloea ramigera biosynthetic thiolase
| |||||||||||
Additional Resources
For additional information, see: Metabolic Disorders
3D structures of Thiolase
References
- ↑ Williams SF, Palmer MA, Peoples OP, Walsh CT, Sinskey AJ, Masamune S. Biosynthetic thiolase from Zoogloea ramigera. Mutagenesis of the putative active-site base Cys-378 to Ser-378 changes the partitioning of the acetyl S-enzyme intermediate. J Biol Chem. 1992 Aug 15;267(23):16041-3. PMID:1353760
- ↑ 2.0 2.1 Kursula P, Ojala J, Lambeir AM, Wierenga RK. The catalytic cycle of biosynthetic thiolase: a conformational journey of an acetyl group through four binding modes and two oxyanion holes. Biochemistry. 2002 Dec 31;41(52):15543-56. PMID:12501183
- ↑ 3.0 3.1 3.2 Merilainen G, Poikela VM, Kursula P, Wierenga RK. The thiolase reaction mechanism: the importance of Asn316 and His348 for stabilizing the enolate intermediate of the Claisen condensation. Biochemistry. 2009 Oct 20. PMID:19842716 doi:10.1021/bi901069h
- ↑ 4.0 4.1 Fukao T, Nguyen HT, Nguyen NT, Vu DC, Can NT, Pham AT, Nguyen KN, Kobayashi H, Hasegawa Y, Bui TP, Niezen-Koning KE, Wanders RJ, de Koning T, Nguyen LT, Yamaguchi S, Kondo N. A common mutation, R208X, identified in Vietnamese patients with mitochondrial acetoacetyl-CoA thiolase (T2) deficiency. Mol Genet Metab. 2010 May;100(1):37-41. Epub 2010 Jan 21. PMID:20156697 doi:10.1016/j.ymgme.2010.01.007
- ↑ Modis Y, Wierenga RK. Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase. J Mol Biol. 2000 Apr 14;297(5):1171-82. PMID:10764581 doi:10.1006/jmbi.2000.3638
- ↑ Haapalainen AM, Merilainen G, Wierenga RK. The thiolase superfamily: condensing enzymes with diverse reaction specificities. Trends Biochem Sci. 2006 Jan;31(1):64-71. Epub 2005 Dec 13. PMID:16356722 doi:10.1016/j.tibs.2005.11.011
- ↑ Jiang C, Kim SY, Suh DY. Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol Phylogenet Evol. 2008 Dec;49(3):691-701. Epub 2008 Sep 12. PMID:18824113 doi:10.1016/j.ympev.2008.09.002
Proteopedia Page Contributors and Editors (what is this?)
Rik Wierenga, Joel L. Sussman, Michal Harel, Satyan Sharma, David Canner