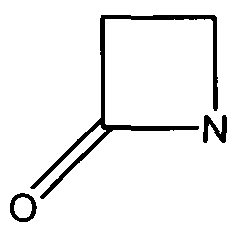
TEM1 Class Antibiotic Resistance Proteins
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='1xpb' size='340' side='right' caption='[[1xpb]] | + | <StructureSection load='1xpb' size='340' side='right' caption='E. coli beta-lactamase Tem-1 complex with sulfate (PDB code[[1xpb]])' scene='72/728117/Highlighted_features_of_tem1-2/1'> |
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1xpb]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1XPB OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1XPB FirstGlance]. <br> | <table><tr><td colspan='2'>[[1xpb]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1XPB OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1XPB FirstGlance]. <br> | ||
| Line 28: | Line 28: | ||
== Function == | == Function == | ||
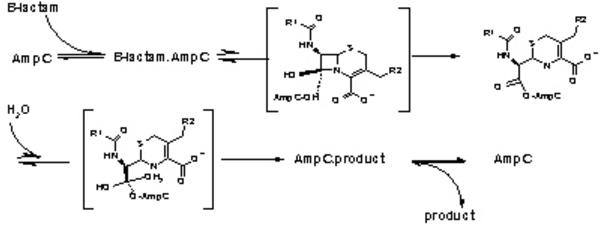
The β-Lactamases has two domains- an alpha helix and a beta sheet of five antiparallel strands, which surround the alpha helix. The catalytic region is known as the oxyanion pocket that occurs between the N-terminus of alpha helix H2 and B3 beta sheet edge. The first step is the acylation of Ser70, one of the catalytic sites. This forms a high-energy acyl-enzyme intermediate, which is then deacylated. This is when the acyl-enzyme is hydrolyzed. The β-Lactam compound has been split and released. The rate determining step is either the acylation or deacylation depending on the antibiotic. However, the opening of the β-Lactam active site increases its chance of inhibition. One of the ways to inhibit β-Lactamases is point mutations in the catalytic region that affect the specificity and catalysis of β-Lactamases enzymes, hindering their activity [4]. | The β-Lactamases has two domains- an alpha helix and a beta sheet of five antiparallel strands, which surround the alpha helix. The catalytic region is known as the oxyanion pocket that occurs between the N-terminus of alpha helix H2 and B3 beta sheet edge. The first step is the acylation of Ser70, one of the catalytic sites. This forms a high-energy acyl-enzyme intermediate, which is then deacylated. This is when the acyl-enzyme is hydrolyzed. The β-Lactam compound has been split and released. The rate determining step is either the acylation or deacylation depending on the antibiotic. However, the opening of the β-Lactam active site increases its chance of inhibition. One of the ways to inhibit β-Lactamases is point mutations in the catalytic region that affect the specificity and catalysis of β-Lactamases enzymes, hindering their activity [4]. | ||
| + | |||
[[Image:Biochem_group_project-function.PNG]] | [[Image:Biochem_group_project-function.PNG]] | ||
| + | |||
Figure 3. The hydrolysis of β-Lactamases where AmpC is the enzyme. There are the three main steps shown with their respective high energy intermediates. (Image Credit: Shoichet Lab in UCSF) | Figure 3. The hydrolysis of β-Lactamases where AmpC is the enzyme. There are the three main steps shown with their respective high energy intermediates. (Image Credit: Shoichet Lab in UCSF) | ||
== Relevance == | == Relevance == | ||
| Line 35: | Line 37: | ||
== Structural highlights == | == Structural highlights == | ||
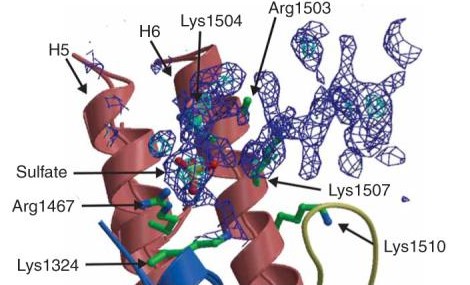
| - | This TEM1 Class Antibiotic Resistant protein is similar in structure to other Beta Lactamases in its secondary structure. The protein consists of | + | This TEM1 Class Antibiotic Resistant protein is similar in structure to other Beta Lactamases in its secondary structure. The protein consists of 263 amino acid residues, which are arranged into a particular structure to provide the function it was designed for. The structure itself consists of two main domains, forming a cleft in between them as the active site for the binding of a sulfate anion ligand. The first domain consists of five beta sheets, with three alpha helices overlaying the side facing the solvent, and another alpha helix flanking the sheets on the adjacent side. The other domain is characterized by its high concentration of alpha helices throughout the secondary structure of this domain. The protein in solution is solvated by 135 water molecules per asymmetric unit. In terms of overall structure, 42.2% consists of alpha helices, 17.5% participate in beta sheets, and 37.2% are turns and coils [8]. Lysine 234, which is present on the wall of the active site as a constituent of one of the beta sheets, has been elucidated as an active binding agent, involved in both the recognition of the substrate and stabilization of the intermediate [8]. Furthermore, the amine group of the Lysine residue is involved in electrostatic binding of the C3 carboxylic group of penicillins. Tyrosine 105 has also been elucidated as a binding mechanism for substrates, as its substitution in the active site has proven to cause decreased catalytic activity [9]. |
== References == | == References == | ||
| Line 46: | Line 48: | ||
4. Fonze, E.; Charlier, P.; To'th, Y.; Vermeire, M.; Raquet, X.; Dubus, A.; Frere, J. M. (1995) TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. ''Acta Crystallogr. D Biol. Crystallogr.'' ''51'', 682-694. | 4. Fonze, E.; Charlier, P.; To'th, Y.; Vermeire, M.; Raquet, X.; Dubus, A.; Frere, J. M. (1995) TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. ''Acta Crystallogr. D Biol. Crystallogr.'' ''51'', 682-694. | ||
| - | 5. Salverda, M. L.; Visser, J. A. G. D.; Barlow, M. Natural Evolution of TEM-1 β-Lactamase: Experimental Reconstruction and Clinical Relevance. FEMS Microbiology Reviews FEMS Microbiol Rev. | + | 5. Salverda, M. L.; Visser, J. A. G. D.; Barlow, M. (2010) Natural Evolution of TEM-1 β-Lactamase: Experimental Reconstruction and Clinical Relevance. ''FEMS Microbiology Reviews FEMS Microbiol Rev.'' ''34'', 1015–1036. |
| - | 6. Neuwirth, C.; Madec, S.; Siebor, E.; Pechinot, A.; Duez, J.-M.; Pruneaux, M.; Fouchereau-Peron, M.; Kazmierczak, A.; Labia, R. TEM-89 Beta -Lactamase Produced by a Proteus Mirabilis Clinical Isolate: New Complex Mutant (CMT 3) with Mutations in Both TEM-59 (IRT-17) and TEM-3. Antimicrobial Agents and Chemotherapy | + | 6. Neuwirth, C.; Madec, S.; Siebor, E.; Pechinot, A.; Duez, J.-M.; Pruneaux, M.; Fouchereau-Peron, M.; Kazmierczak, A.; Labia, R. (2010) TEM-89 Beta -Lactamase Produced by a Proteus Mirabilis Clinical Isolate: New Complex Mutant (CMT 3) with Mutations in Both TEM-59 (IRT-17) and TEM-3. ''Antimicrobial Agents and Chemotherapy'' ''45'', 3591–3594. |
| - | 7. Elander, R. P. Industrial Production of β-Lactam Antibiotics. Appl Microbiol Biotechnol Applied Microbiology and Biotechnology. | + | 7. Elander, R. P. (2003) Industrial Production of β-Lactam Antibiotics. ''Appl Microbiol Biotechnol Applied Microbiology and Biotechnology''. ''61'', 385–392. |
8. Lenfant, F.; Labia, R.; Masson, J. (1991) Replacement of lysine 234 affects transition state stabilization in the active site of ß-lactamase TEM1. ''J. Biol. Chem.'' ''266'', 17187-17194. | 8. Lenfant, F.; Labia, R.; Masson, J. (1991) Replacement of lysine 234 affects transition state stabilization in the active site of ß-lactamase TEM1. ''J. Biol. Chem.'' ''266'', 17187-17194. | ||
9. Doucet, N.; Savard, P. -.; Pelletier, J. N.; Gagné, S. M. (2007) NMR investigation of Tyr105 mutants in TEM-1 ß-lactamase: Dynamics are correlated with function. ''J. Biol. Chem.'' ''282'', 21448-21459. | 9. Doucet, N.; Savard, P. -.; Pelletier, J. N.; Gagné, S. M. (2007) NMR investigation of Tyr105 mutants in TEM-1 ß-lactamase: Dynamics are correlated with function. ''J. Biol. Chem.'' ''282'', 21448-21459. | ||
Current revision
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Kenna Salvatore, Matt O'Malley, Ryan Hunter Wilson, Riley Culhane, Michal Harel