Antizyme Inhibitor
From Proteopedia
(Difference between revisions)
m |
|||
| Line 1: | Line 1: | ||
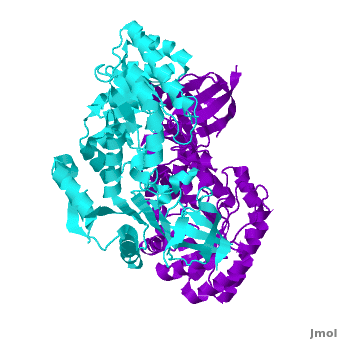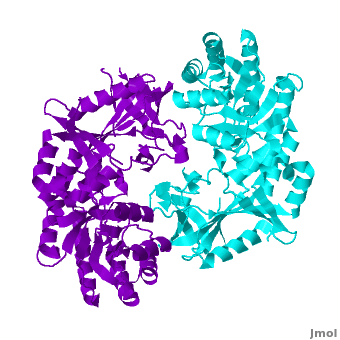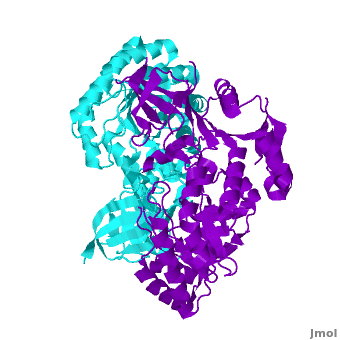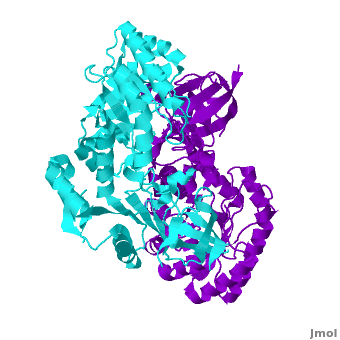
<StructureSection load="" size="400" color="white" caption="Mouse antizyme inhibitor 1 dimer [[3btn]]" scene='Antizyme_Inhibitor/Azi/1' spinBox="true" > | <StructureSection load="" size="400" color="white" caption="Mouse antizyme inhibitor 1 dimer [[3btn]]" scene='Antizyme_Inhibitor/Azi/1' spinBox="true" > | ||
[[Image:AziFig.PNG|left|300px]] | [[Image:AziFig.PNG|left|300px]] | ||
| - | == | + | ==Antizyme Inhibitor== |
{{ABSTRACT_PUBMED_18369191}} | {{ABSTRACT_PUBMED_18369191}} | ||
{{Clear}} | {{Clear}} | ||
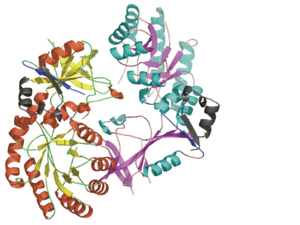
| - | + | Each <scene name='Antizyme_Inhibitor/Monomer/5'>monomer</scene> consists of two domains: a <scene name='Antizyme_Inhibitor/Monomer/6'>TIM-like</scene> α/β-barrel [http://en.wikipedia.org/wiki/TIM_barrel] domain (residues 45–280) and a modified <scene name='Antizyme_Inhibitor/Monomer/7'>Greek key</scene> [http://en.wikipedia.org/wiki/Greek_key] β-sheet domain (residues 8–44 and 281–435). <font color='red'><b>Helices</b></font> [http://en.wikipedia.org/wiki/Alpha_helix] are colored in <font color='red'><b>red</b></font> and <font color='black'><b>β sheets</b></font> [http://en.wikipedia.org/wiki/Beta_sheet] in <font color='black'><b>yellow</b></font>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{{Clear}} | {{Clear}} | ||
A sequence alignment and structural comparison of mouse AzI crystallographic dimer to mouse, human, and [http://en.wikipedia.org/wiki/Trypanosome trypanosome] [http://en.wikipedia.org/wiki/Ornithine_decarboxylase ODC] (mODC, hODC, and tODC, respectively) show high sequence identity (~50%) and structural similarity between AzI and ODC monomers (RMSD values of 1.85 Å, 1.6 Å, and 1.5 Å, respectively). The <scene name='Antizyme_Inhibitor/Azi_odc/10'>structural comparison</scene> of mouse AzI crystallographic dimer (mAzI, <font color='cyan'><b>cyan</b></font> and <font color='blueviolet'><b>blueviolet</b></font>) to mODC (PDB code [[7odc]], (<font color='red'><b>red</b></font> and <font color='lime'><b>lime</b></font>) is shown. Superposition of the <scene name='Antizyme_Inhibitor/Azi_odc/11'>interface</scene> of mAzI and mODC showing the inter-subunit variable loops (AzI residues 355–362 and 387–401). <font color='black'><b>AzI loops</b></font> are in <font color='black'><b>black</b></font>, and <font color='black'><b>ODC loops</b></font> are in <font color='black'><b>yellow</b></font>. | A sequence alignment and structural comparison of mouse AzI crystallographic dimer to mouse, human, and [http://en.wikipedia.org/wiki/Trypanosome trypanosome] [http://en.wikipedia.org/wiki/Ornithine_decarboxylase ODC] (mODC, hODC, and tODC, respectively) show high sequence identity (~50%) and structural similarity between AzI and ODC monomers (RMSD values of 1.85 Å, 1.6 Å, and 1.5 Å, respectively). The <scene name='Antizyme_Inhibitor/Azi_odc/10'>structural comparison</scene> of mouse AzI crystallographic dimer (mAzI, <font color='cyan'><b>cyan</b></font> and <font color='blueviolet'><b>blueviolet</b></font>) to mODC (PDB code [[7odc]], (<font color='red'><b>red</b></font> and <font color='lime'><b>lime</b></font>) is shown. Superposition of the <scene name='Antizyme_Inhibitor/Azi_odc/11'>interface</scene> of mAzI and mODC showing the inter-subunit variable loops (AzI residues 355–362 and 387–401). <font color='black'><b>AzI loops</b></font> are in <font color='black'><b>black</b></font>, and <font color='black'><b>ODC loops</b></font> are in <font color='black'><b>yellow</b></font>. | ||
Revision as of 08:11, 19 March 2019
| |||||||||||
3D structures of Antizyme Inhibitor
Updated on 19-March-2019
3btn – AzI – mouse
1zo0 – AzI – rat - NMR
4zgz – AzI 1 + ornithine decarboxylase antizyme 1 – human
Additional Resources
For additional information, see: Cancer
Reference
Shira Albeck, Orly Dym, Tamar Unger, Zohar Snapir, Zippy Bercovich and Chaim Kahana. Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function. Protein Sci. 2008 May; 17(5): 793-802. Epub 2008 Mar 27.
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Michal Harel, Joel L. Sussman, Dinesh Kulhary, David Canner, Jaime Prilusky, Orly Dym