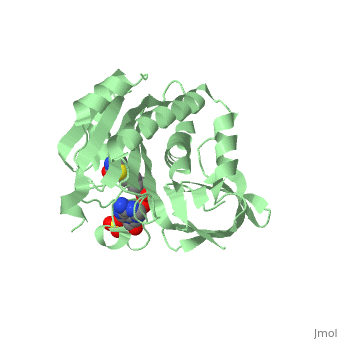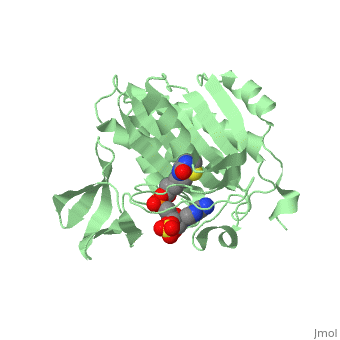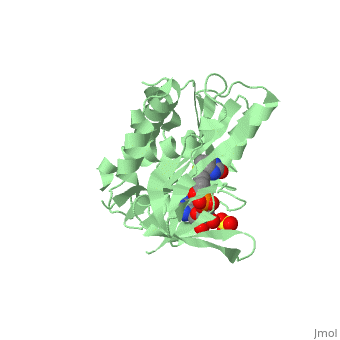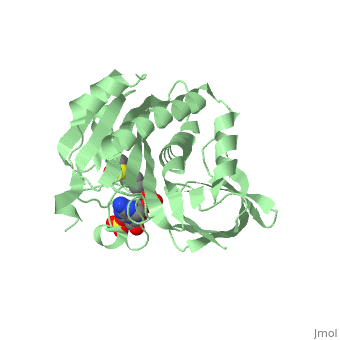Biotin Protein Ligase
From Proteopedia
(Difference between revisions)
| (23 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='4op0' size='350' side='right' scene= caption='Biotin protein ligase complex with biotinyl-5-AMP and sulfate (PDB code [[4op0]])'> | |
| - | The attachment of [http://en.wikipedia.org/wiki/Biotin biotin] onto requiring proteins is catalysed by the ubiquitous enzyme '''biotin protein ligase''' (BPL), also known as the biotin inducible repressor, BirA, in E.coli and holocarboxylase synthase (HCS) in mammals. It was once believed a separate HCS existed for each of the carboxylases. However, with the availability of modern recombinant DNA technology and complete genome sequences, there is good evidence that only one biotin protein ligase is present in most bacteria, yeast and mammals. Arabidopsis thaliana and other plants species are a notable exception to this rule as they contain two HCS genes, one encoding a cytoplasmic enzyme and the other a chloroplast targeted enzyme. | + | __TOC__ |
| + | == Function == | ||
| + | |||
| + | The attachment of [http://en.wikipedia.org/wiki/Biotin biotin] onto requiring proteins is catalysed by the ubiquitous enzyme '''biotin protein ligase''' (BPL), also known as the biotin inducible repressor, BirA, in ''E.coli'' and holocarboxylase synthase (HCS) in mammals. It was once believed a separate HCS existed for each of the carboxylases. However, with the availability of modern recombinant DNA technology and complete genome sequences, there is good evidence that only one biotin protein ligase is present in most bacteria, yeast and mammals. ''Arabidopsis thaliana'' and other plants species are a notable exception to this rule as they contain two HCS genes, one encoding a cytoplasmic enzyme and the other a chloroplast targeted enzyme. | ||
Biotinylation is catalysed through a two-step reaction where biotin is first activated to biotinyl-5′-AMP in an ATP dependent manner. The biotin is then transferred onto the ε-amino group of a specific target lysine residue. The reaction mechanism is related to that of amino acyl-tRNA synthetases and lipoyl ligases where the reaction proceeds through the formation of an adenylated intermediate, suggesting a common ancestral relationship <ref>pmid 18442489</ref> . | Biotinylation is catalysed through a two-step reaction where biotin is first activated to biotinyl-5′-AMP in an ATP dependent manner. The biotin is then transferred onto the ε-amino group of a specific target lysine residue. The reaction mechanism is related to that of amino acyl-tRNA synthetases and lipoyl ligases where the reaction proceeds through the formation of an adenylated intermediate, suggesting a common ancestral relationship <ref>pmid 18442489</ref> . | ||
| - | Of all the BPL’s, | + | Of all the BPL’s, '''BirA bifunctional protein''' is by far the most characterised and understood family member. BirA catalyzes the two activities of post-translational biotinylation and repression of transcription initiation<ref>pmid 28466579</ref>. A recent ensemble of BPL structures from the thermophilic archea Pirococcus Horikoshii OT3 <ref>pmid 16510991</ref> have also provided new insights into the catalytic mechanism of BPLs. |
| - | BirA (35.5 kDa) contains three distinct domains that have been determined at 2.3 Å resolution in 1992 through X-ray crystallography in the unliganed form, <scene name='Biotin_Protein_Ligase/Apo_ecbpl/1'>apo_EcBPL</scene> . The monomeric structure measures 75 Å x 35 Å x 30 Å for the unliganded "apo" structure <ref>pmid 1409631</ref>. The <scene name='Biotin_Protein_Ligase/Apo_ecbpl_nterm/3'>N-terminal</scene> 22-46 residues adopt a helix-turn-helix motif, a structure associated with DNA binding proteins. <font color="purple"><scene name='Biotin_Protein_Ligase/Apo_ecbpl_cat/1'>The central domain </scene></font> consists of five α helices, 7 strands of mixed β-sheets as well as four poorly-defined loops that appear in pairs in the 3D structure. These loops consist of residues 110-128, 212-233 and 140 146 and 193-199. The <scene name='Biotin_Protein_Ligase/Apo_ecbpl_cterm/2'>C- terminus</scene> consists of 6 strands which form a β-sandwich that seals the end of the enzyme and has been found to function in the transfer of biotin onto BCCP. | + | == Structural highlights == |
| - | Upon biotin binding, the protein homodimerises and the unstructured loops become more ordered. | + | |
| + | BirA (35.5 kDa) contains three distinct domains that have been determined at 2.3 Å resolution in 1992 through X-ray crystallography in the unliganed form, <scene name='Biotin_Protein_Ligase/Apo_ecbpl/1'>apo_EcBPL</scene> . The monomeric structure measures 75 Å x 35 Å x 30 Å for the unliganded "apo" structure <ref>pmid 1409631</ref>. The <scene name='Biotin_Protein_Ligase/Apo_ecbpl_nterm/3'>N-terminal</scene> 22-46 residues adopt a helix-turn-helix motif, a structure associated with DNA binding proteins. <font color="purple"><scene name='Biotin_Protein_Ligase/Apo_ecbpl_cat/1'>The central domain </scene></font> consists of five α helices, 7 strands of mixed β-sheets as well as four poorly-defined loops that appear in pairs in the 3D structure. These loops consist of residues 110-128, 212-233 and 140 146 and 193-199. The <scene name='Biotin_Protein_Ligase/Apo_ecbpl_cterm/2'>C- terminus</scene> consists of 6 strands which form a β-sandwich that seals the end of the enzyme and has been found to function in the transfer of biotin onto BCCP. Upon biotin binding, the protein homodimerises and the unstructured loops become more ordered. <scene name='23/237702/Cv/5'>The binding pocket of biotinyl-5'-AMP to BirA is shown here</scene>. | ||
| + | *<scene name='23/237702/Cv/4'>Biotinyl-5'-AMP interactions with BirA</scene>. | ||
==Additional Resources== | ==Additional Resources== | ||
| - | For additional information, see: [[Carbohydrate Metabolism]] | + | For additional information, see: [[Carbohydrate Metabolism]]; [[Ligases]]. |
<br /> | <br /> | ||
==3D structures of Biotin Protein Ligase== | ==3D structures of Biotin Protein Ligase== | ||
| + | [[Biotin Protein Ligase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==References== | ==References== | ||
Current revision
| |||||||||||
References
- ↑ Pendini NR, Bailey LM, Booker GW, Wilce MC, Wallace JC, Polyak SW. Microbial biotin protein ligases aid in understanding holocarboxylase synthetase deficiency. Biochim Biophys Acta. 2008 Jul-Aug;1784(7-8):973-82. Epub 2008 Apr 9. PMID:18442489 doi:10.1016/j.bbapap.2008.03.011
- ↑ Wang J, Beckett D. A conserved regulatory mechanism in bifunctional biotin protein ligases. Protein Sci. 2017 Aug;26(8):1564-1573. doi: 10.1002/pro.3182. Epub 2017 May 11. PMID:28466579 doi:http://dx.doi.org/10.1002/pro.3182
- ↑ Bagautdinov B, Kuroishi C, Sugahara M, Kunishima N. Purification, crystallization and preliminary crystallographic analysis of the biotin-protein ligase from Pyrococcus horikoshii OT3. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005 Feb 1;61(Pt, 2):193-5. Epub 2005 Jan 8. PMID:16510991 doi:10.1107/S1744309104034360
- ↑ Wilson KP, Shewchuk LM, Brennan RG, Otsuka AJ, Matthews BW. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9257-61. PMID:1409631
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Nicole R Pendini, Alexander Berchansky, David Canner, Jaime Prilusky