Calcium uptake protein 1
From Proteopedia
(Difference between revisions)
| (16 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | ==Structure== | + | ==Structure of MICU1== |
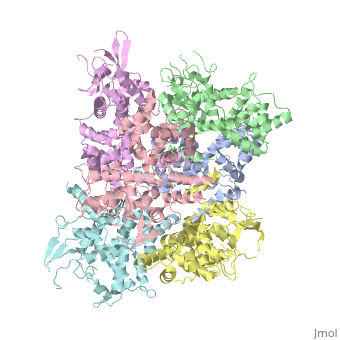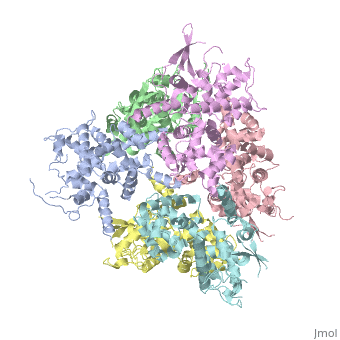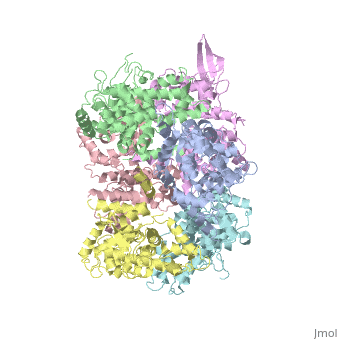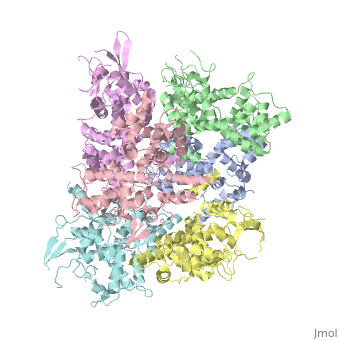
| - | <StructureSection load='4NSC' size='340' side=' | + | <StructureSection load='4NSC' size='340' side='left' caption='Human mitochondrial calcium uptake protein 1 (PDB code [[4nsc]])' scene=''> |
| - | MICU 1 (Mitochondrial Calcium Uptake 1) is a key regulator of mitochondrial calcium uniporter (MCU) required to increase calcium uptake by MCU when cytoplasmic calcium is high. | + | '''MICU 1''' ('''Mitochondrial Calcium Uptake Protein 1''') is a key regulator of mitochondrial calcium uniporter (MCU) required to increase calcium uptake by MCU when cytoplasmic calcium is high. |
It also regulates glucose-dependent insulin secretion in pancreatic beta-cells by regulating mitochondrial calcium uptake. | It also regulates glucose-dependent insulin secretion in pancreatic beta-cells by regulating mitochondrial calcium uptake. | ||
| + | |||
| + | <table><tr><td colspan='2'> | ||
| + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat">Ca 2+</td></tr> | ||
| + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">MICU 2, MCU, EMRE</td></tr> | ||
| + | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">CBARA, chromosome 10 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 Homo sapiens])</td></tr> | ||
| + | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat">Regulator of Ca 2+ uptake in the mitochondria</td></tr> | ||
| + | </table> | ||
| + | |||
| + | == History == | ||
| + | MICU1 was identified as an essential element of mitochondrial calcium uptake in 2010. It was crystallise in the presence of calcium chloride and methyl-pentanediol (MPD). Recent discover of this protein doesn’t permit to have more information about its evolutionary conservation. | ||
== Function == | == Function == | ||
| Line 10: | Line 20: | ||
When it is not linked to Ca2+ it has an hexameric conformation. In this conformation MICU 1 silences the activity of MCU/EMRE and stop the Ca 2+ from entering the mitochondria by blocking the entrance of the channel. | When it is not linked to Ca2+ it has an hexameric conformation. In this conformation MICU 1 silences the activity of MCU/EMRE and stop the Ca 2+ from entering the mitochondria by blocking the entrance of the channel. | ||
When there is a lot of calcium in the intermembrane so in the cytoplasm, Ca2+ binds to MICU1, then it changes its conformation, separates into multiple homooligomers that results in opens the “gate” in front of the MCU channel, letting the calcium enter into the matrix. | When there is a lot of calcium in the intermembrane so in the cytoplasm, Ca2+ binds to MICU1, then it changes its conformation, separates into multiple homooligomers that results in opens the “gate” in front of the MCU channel, letting the calcium enter into the matrix. | ||
| - | |||
| - | |||
| - | == Disease == | ||
| - | If MICU1 gene is modified, Ca2+ can be load in higher concentration in the mitochondry resulting Myopathy with extrapyramidal signs (MPXPS). This is an autosomal recessive disorder characterized by early-onset proximal muscle weakness with a static course and moderately to grossly elevated serum creatine kinase levels accompanied by learning difficulties. Most patients develop subtle extrapyramidal motor signs that progress to a debilitating disorder of involuntary movement with variable features, including chorea, tremor, dystonic posturing and orofacial dyskinesia. Additional variable features include ataxia, microcephaly, ophthalmoplegia, ptosis, optic atrophy and axonal peripheral neuropathy. | ||
== Structural highlights == | == Structural highlights == | ||
| + | MICU 1 is a ~54kDa transmembrane protein composed by an amino-terminal mitochondrial targeting sequence, a transmembrane helix and a cytosolic C-terminus which contain different important domains describe after. | ||
| + | |||
In absence of Ca2+, MICU1 is an homohexamer with a <scene name='72/723172/Secondary_structure_of_micu_1/1'>secondary structure</scene> composed by several alpha helix, beta sheets and loops. When it <scene name='72/723172/Protein_dimer_when_ca_is_bound/3'>links to this ion</scene> , it becomes a homooligomer. | In absence of Ca2+, MICU1 is an homohexamer with a <scene name='72/723172/Secondary_structure_of_micu_1/1'>secondary structure</scene> composed by several alpha helix, beta sheets and loops. When it <scene name='72/723172/Protein_dimer_when_ca_is_bound/3'>links to this ion</scene> , it becomes a homooligomer. | ||
MICU 1 is composed by differents domains : | MICU 1 is composed by differents domains : | ||
| - | :- C helix region (or Coiled-coil domain) : this domain permits different interactions with other proteins (MCU/MICU2) and is required to assemble free Ca2+ with homohexamer. | + | :- <scene name='72/723172/C-helix_region/3'>C-helix region</scene> (or Coiled-coil domain) : this domain permits different interactions with other proteins (MCU/MICU2) and is required to assemble free Ca2+ with homohexamer. |
| - | :- Polybasic region : it’s a domain which permits protein-protein interactions | + | :- <scene name='72/723172/Polybasic_region/1'>Polybasic region</scene> : it’s a domain which permits protein-protein interactions |
:- <scene name='72/723172/Ef_domains/1'>EF-hand domains</scene> There are 2 of them. They are the most important domains in MICU1 protein because they contain the Ca2+ binding region and detect the concentration of calcium in the cell for activation of signalling. An EF-hand domain is a calcium sensor. An EF-hand domain is a helix loop helix structural domain that means that it is 2 alpha helices linked by a short loop region. Ca2+ ions are coordinated in this space thanks to ligands within the loop : in EF-1 residues concerning are Asp231, Asn233, Asp235, Glu237 and Glu242 and in EF-2 they are Asp421, Asp423, Asn425, Glu427 and Glu432. When Ca2+ binds itself to this domain, the protein changes its conformation to expose a domain that can interact with other proteins. | :- <scene name='72/723172/Ef_domains/1'>EF-hand domains</scene> There are 2 of them. They are the most important domains in MICU1 protein because they contain the Ca2+ binding region and detect the concentration of calcium in the cell for activation of signalling. An EF-hand domain is a calcium sensor. An EF-hand domain is a helix loop helix structural domain that means that it is 2 alpha helices linked by a short loop region. Ca2+ ions are coordinated in this space thanks to ligands within the loop : in EF-1 residues concerning are Asp231, Asn233, Asp235, Glu237 and Glu242 and in EF-2 they are Asp421, Asp423, Asn425, Glu427 and Glu432. When Ca2+ binds itself to this domain, the protein changes its conformation to expose a domain that can interact with other proteins. | ||
| + | |||
| + | == Disease == | ||
| + | If MICU1 gene is modified, Ca2+ can be load in higher concentration in the mitochondry resulting Myopathy with extrapyramidal signs (MPXPS). This is an autosomal recessive disorder characterized by early-onset proximal muscle weakness with a static course and moderately to grossly elevated serum creatine kinase levels accompanied by learning difficulties. Most patients develop subtle extrapyramidal motor signs that progress to a debilitating disorder of involuntary movement with variable features, including chorea, tremor, dystonic posturing and orofacial dyskinesia. Additional variable features include ataxia, microcephaly, ophthalmoplegia, ptosis, optic atrophy and axonal peripheral neuropathy. | ||
| + | |||
| + | </StructureSection> | ||
| + | |||
| + | == 3D Structures of calcium uptake protein 1 == | ||
| + | |||
| + | [[Calcium uptake protein 3D structures]] | ||
== See Also == | == See Also == | ||
| Line 29: | Line 46: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | :https://www.researchgate.net/publication/260152737_Structural_and_mechanistic_insights_into_MICU1_regulation_of_mitochondrial_calcium_uptake | ||
| + | :http://ghr.nlm.nih.gov/gene/MICU1 | ||
| + | :http://www.rcsb.org/pdb/home/home.do | ||
| + | :http://www.uniprot.org/uniprot/Q9BPX6 | ||
Current revision
Contents |
Structure of MICU1
| |||||||||||
3D Structures of calcium uptake protein 1
Calcium uptake protein 3D structures
See Also
References
- https://www.researchgate.net/publication/260152737_Structural_and_mechanistic_insights_into_MICU1_regulation_of_mitochondrial_calcium_uptake
- http://ghr.nlm.nih.gov/gene/MICU1
- http://www.rcsb.org/pdb/home/home.do
- http://www.uniprot.org/uniprot/Q9BPX6