C-di-GMP specific phosphodiesterases
From Proteopedia
(Difference between revisions)
| (10 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='2w27' size=' | + | <StructureSection load='2w27' size='350' side='right' scene='' caption='C-di-GMP specific phosphodiesterase complex with guanosine-5-monophosphate (stick model) and Ca+2 ions (green) (PDB code [[2w27]])'> |
| - | + | == YkuI == | |
| - | + | ||
| - | == | + | |
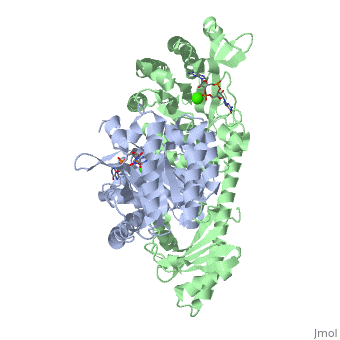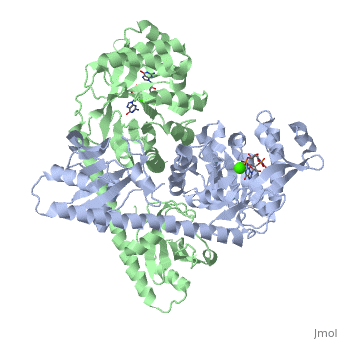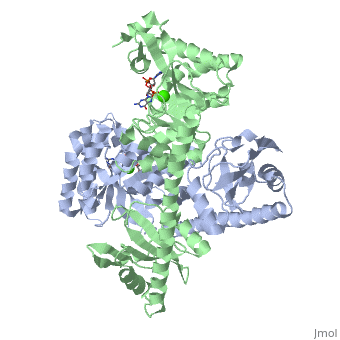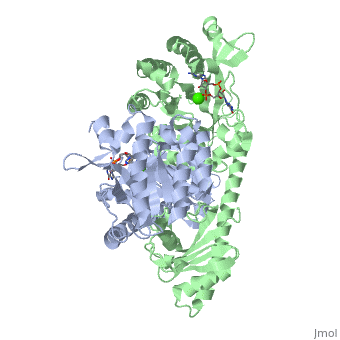
<scene name='User:Tilman_Schirmer/Sandbox_202/Yuki_dimer/3'>YukI</scene> is a dimer with each protomer composed of an <scene name='User:Tilman_Schirmer/Sandbox_202/Eal_domain/1'>EAL domain</scene> (with TIM barrel fold), a long <scene name='User:Tilman_Schirmer/Sandbox_202/Connection/1'>helical linker</scene>, and a <scene name='User:Tilman_Schirmer/Sandbox_202/Pas_like_domain/1'>PAS-like domain</scene>. <br> | <scene name='User:Tilman_Schirmer/Sandbox_202/Yuki_dimer/3'>YukI</scene> is a dimer with each protomer composed of an <scene name='User:Tilman_Schirmer/Sandbox_202/Eal_domain/1'>EAL domain</scene> (with TIM barrel fold), a long <scene name='User:Tilman_Schirmer/Sandbox_202/Connection/1'>helical linker</scene>, and a <scene name='User:Tilman_Schirmer/Sandbox_202/Pas_like_domain/1'>PAS-like domain</scene>. <br> | ||
| Line 8: | Line 6: | ||
The <scene name='User:Tilman_Schirmer/Sandbox_202/Active_site_close/4'>active site </scene> is situated at the C-terminal end of the central β-barrel of the EAL domain. The residues of the active site <scene name='User:Tilman_Schirmer/Sandbox_202/Active_site_close_labels/1'>(here shown with labels)</scene> are well conserved, including E33 and L35 of the EAL signature motif. <br> | The <scene name='User:Tilman_Schirmer/Sandbox_202/Active_site_close/4'>active site </scene> is situated at the C-terminal end of the central β-barrel of the EAL domain. The residues of the active site <scene name='User:Tilman_Schirmer/Sandbox_202/Active_site_close_labels/1'>(here shown with labels)</scene> are well conserved, including E33 and L35 of the EAL signature motif. <br> | ||
| - | The <scene name='User:Tilman_Schirmer/Sandbox_202/Active_site_ligand/3'>c-di-GMP substrate</scene> is bound flat upon the actve site with the divalent metal (here Ca<sup>++</sup>) being sandwiched between binding site and | + | The <scene name='User:Tilman_Schirmer/Sandbox_202/Active_site_ligand/3'>c-di-GMP substrate</scene> is bound flat upon the actve site with the divalent metal (here Ca<sup>++</sup>) being sandwiched between binding site and [[C-di-GMP]]. |
== BlrP1 == | == BlrP1 == | ||
| Line 27: | Line 25: | ||
<scene name='User:Tilman_Schirmer/Sandbox_202/Blrp1_active_site_ligand_hoh/2'>Active site + ligand + Mn + HOH</scene> | <scene name='User:Tilman_Schirmer/Sandbox_202/Blrp1_active_site_ligand_hoh/2'>Active site + ligand + Mn + HOH</scene> | ||
| + | |||
| + | == Additional Resources == | ||
| + | [[C-di-GMP signaling|See also C-di-GMP signaling]] | ||
</StructureSection> | </StructureSection> | ||
| - | + | ==3D structure of C-di-GMP phosphodiesterase== | |
| - | ==3D structure of | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
[[Phosphodiesterase]] | [[Phosphodiesterase]] | ||
Current revision
| |||||||||||
3D structure of C-di-GMP phosphodiesterase
References
YkuI structure 2w27:
- Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Basle A, Massa C, Collart FR, Schirmer T, Anderson WF. Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J Biol Chem. 2009 May 8;284(19):13174-84. Epub 2009 Feb 24. PMID:19244251 doi:10.1074/jbc.M808221200
BlrP1 structure 3gg0:
- Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009 Jun 18;459(7249):1015-8. PMID:19536266 doi:10.1038/nature07966
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Joel L. Sussman, Alexander Berchansky, Tilman Schirmer