We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Formate dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
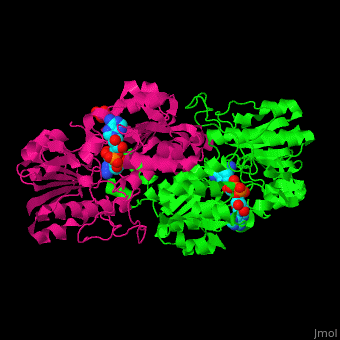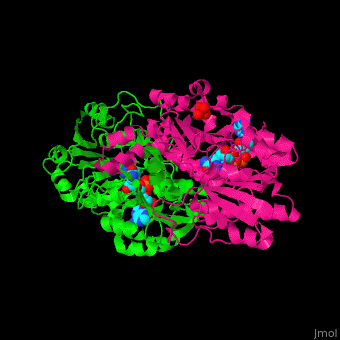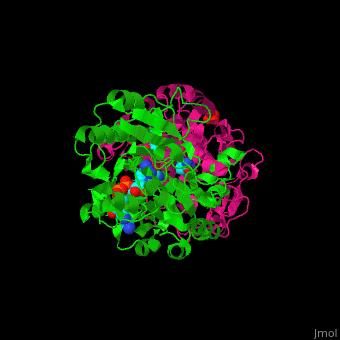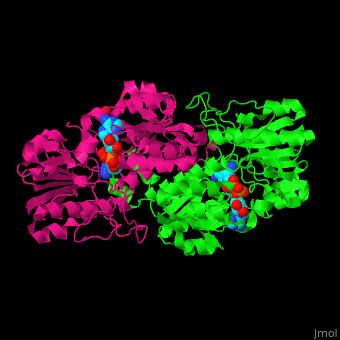
*<scene name='49/491980/Cv/8'>NAD binding site</scene> in NAD-dependent formate dehydrogenase ([[2nad]]). Water molecules shown as red spheres. | *<scene name='49/491980/Cv/8'>NAD binding site</scene> in NAD-dependent formate dehydrogenase ([[2nad]]). Water molecules shown as red spheres. | ||
*<scene name='49/491980/Cv/9'>Azide ion binding site</scene>. | *<scene name='49/491980/Cv/9'>Azide ion binding site</scene>. | ||
| + | |||
| + | ==3D structure of formate dehydrogenase== | ||
| + | [[Formate dehydrogenase 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
==3D structure of formate dehydrogenase== | ==3D structure of formate dehydrogenase== | ||
| Line 22: | Line 26: | ||
**[[3n7u]] - ArFDH + NAD + N3<BR /> | **[[3n7u]] - ArFDH + NAD + N3<BR /> | ||
**[[5dn9]] - CbFDH + NAD + N3<BR /> | **[[5dn9]] - CbFDH + NAD + N3<BR /> | ||
| + | **[[6d4b]], [[6d4c]] – CbFDH (mutant) + NAD + N3<br /> | ||
**[[3wr5]] – FDH + NAD – ''Thiobacillus''<br /> | **[[3wr5]] – FDH + NAD – ''Thiobacillus''<br /> | ||
**[[4xyg]] – GmFDH – ''Granulicella mallensis''<br /> | **[[4xyg]] – GmFDH – ''Granulicella mallensis''<br /> | ||
**[[4xyb]] – GmFDH + NADP <br /> | **[[4xyb]] – GmFDH + NADP <br /> | ||
**[[4xye]] – GmFDH + NAD<br /> | **[[4xye]] – GmFDH + NAD<br /> | ||
| + | **[[1qp8]] – FDH + NADP – ''Pyrobaculum aerophilum''<br /> | ||
*Mo-dependent FDH | *Mo-dependent FDH | ||
Revision as of 09:26, 3 July 2019
| |||||||||||
3D structure of formate dehydrogenase
Updated on 03-July-2019