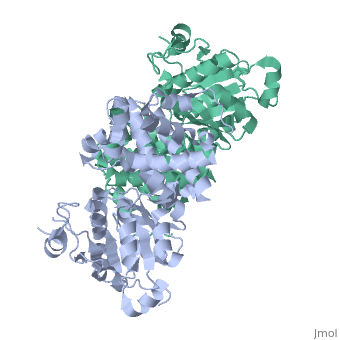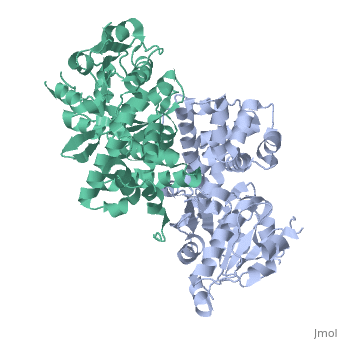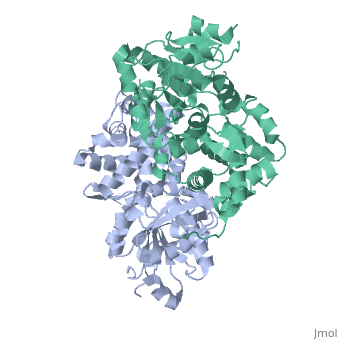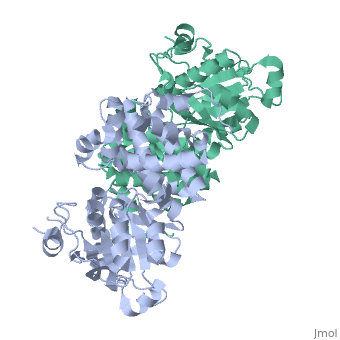
Glycerol-3-Phosphate Dehydrogenase
From Proteopedia
(Difference between revisions)
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1yj8' size='350' side='right' scene='' caption='Glycerol-3-phosphate dehydrogenase trimer [[1yj8]]'> | |
| - | + | ||
'''Glycerol-3-phosphate dehydrogenase''' (GlpD) is a membrane bound enzyme in prokaryotes and in eukaryotes. Glycerol 3-Phosphate Dehydrogenase (GlpD) is an oxidoreductase enzyme which catalyzes the reduction in [http://en.wikipedia.org/wiki/File:Dihydroxyacetone_phosphate_to_glycerol_3-phosphate_en.svg reaction] of Dihydroxyacetone Phosphate to Glycerol 3-Phosphate, with NADH as the reductant. GlpD is involved in many cellular functions such as phospholipids biosynthesis, respiration and metabolism. <ref name="cellfunction">PMID:18296637</ref>The GlpD is a dimer consisting of two subunits which contain the catabolite activator protein (CAP)-Domain,the flavin adenine dinucleotide(FAD)-Domain and a ubiquinone analogue, MD. | '''Glycerol-3-phosphate dehydrogenase''' (GlpD) is a membrane bound enzyme in prokaryotes and in eukaryotes. Glycerol 3-Phosphate Dehydrogenase (GlpD) is an oxidoreductase enzyme which catalyzes the reduction in [http://en.wikipedia.org/wiki/File:Dihydroxyacetone_phosphate_to_glycerol_3-phosphate_en.svg reaction] of Dihydroxyacetone Phosphate to Glycerol 3-Phosphate, with NADH as the reductant. GlpD is involved in many cellular functions such as phospholipids biosynthesis, respiration and metabolism. <ref name="cellfunction">PMID:18296637</ref>The GlpD is a dimer consisting of two subunits which contain the catabolite activator protein (CAP)-Domain,the flavin adenine dinucleotide(FAD)-Domain and a ubiquinone analogue, MD. | ||
| Line 6: | Line 5: | ||
===Structure=== | ===Structure=== | ||
| - | [[Image:FINAL.png|thumb|Glycerol 3-Phosphate Dehydrogenase]] | + | [[Image:FINAL.png|left|thumb|Glycerol 3-Phosphate Dehydrogenase]] |
| - | + | {{Clear}} | |
GlpD is a dimer that consists of two subunits; α and β. The GlpD structure also contains seven ligands; 1,3-Dihydroxyacetonephosphate (13P), β-Octylglucoside (βOG), 1,2-Ethanediol (EDO), Flavin-Adenine Dinucleotide (FAD), Imidazole (IMD), PO4 (Phosphate Ion) and N-(Tris(Hydroxymethyl)methyl)-3-Aminopropanesulfonic Acid (T3A). The active sites on GlpD are the Cap-Domain, FAD- Domain and a ubiquinone substrate analogue, menadione (MD). | GlpD is a dimer that consists of two subunits; α and β. The GlpD structure also contains seven ligands; 1,3-Dihydroxyacetonephosphate (13P), β-Octylglucoside (βOG), 1,2-Ethanediol (EDO), Flavin-Adenine Dinucleotide (FAD), Imidazole (IMD), PO4 (Phosphate Ion) and N-(Tris(Hydroxymethyl)methyl)-3-Aminopropanesulfonic Acid (T3A). The active sites on GlpD are the Cap-Domain, FAD- Domain and a ubiquinone substrate analogue, menadione (MD). | ||
| Line 16: | Line 15: | ||
<scene name='Sandbox_189/Fad/2'>FAD Active Site</scene> | <scene name='Sandbox_189/Fad/2'>FAD Active Site</scene> | ||
| - | The N-terminal FAD-Domain exists in each monomer subunit of GlpD and is embedded into the phospholipid membrane bilayer.Substrate binding occurs at this domain which causes a conformational change to the structure of the GlpD enzyme. The FAD-domain plays a major role in metabolism and energy synthesis. | + | The N-terminal FAD-Domain exists in each monomer subunit of GlpD and is embedded into the phospholipid membrane bilayer.Substrate binding occurs at this domain which causes a conformational change to the structure of the GlpD enzyme. The FAD-domain plays a major role in metabolism and energy synthesis. The active site is found in a cleft between the two domains<ref>PMID:14717590</ref>. |
===Function=== | ===Function=== | ||
| - | GlpD functions in the intracellular membrane of E. coli and in the inner-mitochondrial membrane of eukaryotes. In E. Coli, GlpD catalyzes and reduces the reaction of dihydroxyacetone phosphate to glycerol 3-phosphate in the [http://www.pnas.org/content/105/9/3280/F1.large.jpg glycerol metabolism pathway]. The binding of the substrate analogues (glyceraldehydes 3-phosphate, glyceric acid 2-phosphate and phosphoenolpyruvate, dihydroxyacetone phosphate)or UQ substrate analogues (2-n-heptyl-4-hydroxyquinoline N-oxide and menadione). The conformational change of the structure and resiudes of GlpD catalyzes many different metabolic reactions. | + | GlpD functions in the intracellular membrane of E. coli and in the inner-mitochondrial membrane of eukaryotes. In E. Coli, GlpD catalyzes and reduces the reaction of dihydroxyacetone phosphate (DHAP) to glycerol 3-phosphate in the [http://www.pnas.org/content/105/9/3280/F1.large.jpg glycerol metabolism pathway]. The binding of the substrate analogues (glyceraldehydes 3-phosphate, glyceric acid 2-phosphate and phosphoenolpyruvate, dihydroxyacetone phosphate) or UQ substrate analogues (2-n-heptyl-4-hydroxyquinoline N-oxide and menadione). The conformational change of the structure and resiudes of GlpD catalyzes many different metabolic reactions. |
===Metabolic Pathways=== | ===Metabolic Pathways=== | ||
| Line 26: | Line 25: | ||
====Phosphoplipid Biosynthesis==== | ====Phosphoplipid Biosynthesis==== | ||
| - | GlpD reduces | + | GlpD reduces DHAP to glycerol 3-phosphate. Then the glycerol 3-phosphate is catalyzed by acyl transferase to 1-acylglyverol-3-phosphate, and then another acyl transferase catalyzes that to a phosphatidic acid. head groups are added to the phosphatidic acid to synthesize phospholipids. |
====Glyceroneogenesis==== | ====Glyceroneogenesis==== | ||
| Line 38: | Line 37: | ||
===Diseases=== | ===Diseases=== | ||
| - | GlpD is involved in diseases such as | + | GlpD is involved in diseases such as Alzeheimer`s, muscle dystrophy, hyaline membrane diseases and many more. |
==3D structures of glycerol-3-phosphate dehydrogenase== | ==3D structures of glycerol-3-phosphate dehydrogenase== | ||
| + | [[Glycerol-3-phosphate dehydrogenase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ==References== | |
<references /> | <references /> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Yeh JI, Chinte U, Du S. Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. Proc Natl Acad Sci U S A. 2008 Mar 4;105(9):3280-5. Epub 2008 Feb 22. PMID:18296637
- ↑ Yeh JI, Charrier V, Paulo J, Hou L, Darbon E, Claiborne A, Hol WG, Deutscher J. Structures of enterococcal glycerol kinase in the absence and presence of glycerol: correlation of conformation to substrate binding and a mechanism of activation by phosphorylation. Biochemistry. 2004 Jan 20;43(2):362-73. PMID:14717590 doi:10.1021/bi034258o
Proteopedia Page Contributors and Editors (what is this?)
Indu Toora, Michal Harel, Alexander Berchansky, David Canner, Andrea Gorrell, Andrew Rebeyka, Jaime Prilusky