Mannosidase
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Structural highlights == | == Structural highlights == | ||
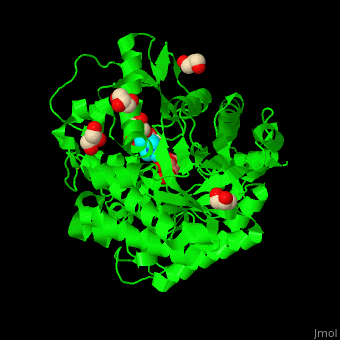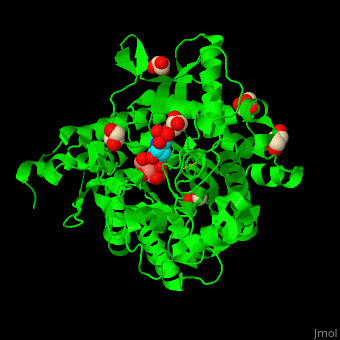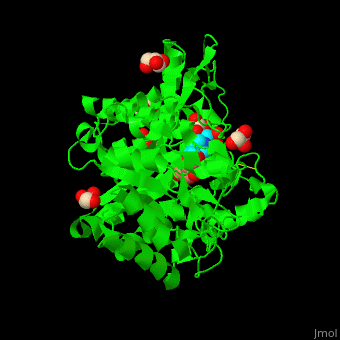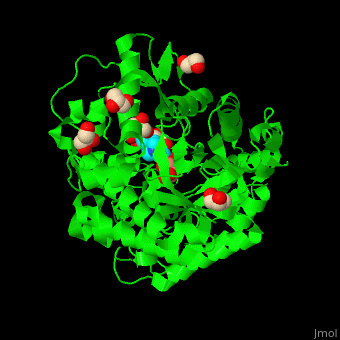
<scene name='45/458458/Cv/5'>The active site of β-MAN contains β-mannose</scene><ref>PMID:24100330</ref>. Water molecules shown as red spheres. | <scene name='45/458458/Cv/5'>The active site of β-MAN contains β-mannose</scene><ref>PMID:24100330</ref>. Water molecules shown as red spheres. | ||
| + | |||
| + | ==3D structures of mannosidase== | ||
| + | [[Mannosidase 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
==3D structures of mannosidase== | ==3D structures of mannosidase== | ||
| Line 17: | Line 21: | ||
*α-MAN | *α-MAN | ||
| - | **[[1hcu]] - TrαMAN – ''Trichoderma reesei''<br /> | ||
**[[1o7d]] - αMAN – bovine<br /> | **[[1o7d]] - αMAN – bovine<br /> | ||
| - | **[[2wvy]], [[2ww2]], [[4acy]], [[4acz]], [[4ad0]], [[4c1s]] - αMAN - ''Bacterioides thetaiotaomicron'' | + | **[[6b9o]] - jbMAN – jack bean<br /> |
| + | **[[2wvy]], [[2ww2]], [[4acy]], [[4acz]], [[4ad0]], [[4c1s]] - αMAN - ''Bacterioides thetaiotaomicron''<br /> | ||
| + | **[[1hcu]] - TrαMAN – ''Trichoderma reesei''<br /> | ||
*α-MAN class I | *α-MAN class I | ||
| Line 26: | Line 31: | ||
**[[1fo2]] - hαMAN I + deoxymannojirimycin<br /> | **[[1fo2]] - hαMAN I + deoxymannojirimycin<br /> | ||
**[[1fo3]] - hαMAN I+ kifunensine<br /> | **[[1fo3]] - hαMAN I+ kifunensine<br /> | ||
| + | **[[1x9d]] - hαMAN I residues 243-699 + polysaccharide analog<br /> | ||
| + | **[[1nxc]] - mαMAN I + mannose – mouse<br /> | ||
| + | **[[6b9p]] - jbMAN + inhibitor<br /> | ||
**[[5jm0]] - yαMAN I – yeast<br /> | **[[5jm0]] - yαMAN I – yeast<br /> | ||
| - | **[[1dl2]] - yαMAN I catalytic domain + mannose - | + | **[[1dl2]] - yαMAN I catalytic domain + mannose <br /> |
| + | **[[1g6i]] - yαMAN I + deoxymannojirimycin<br /> | ||
**[[2ri8]] - PcαMAN I + glycerol – ''Penicillium citrinum''<br /> | **[[2ri8]] - PcαMAN I + glycerol – ''Penicillium citrinum''<br /> | ||
**[[1kre]], [[1kkt]] - PcαMAN I + mannose<br /> | **[[1kre]], [[1kkt]] - PcαMAN I + mannose<br /> | ||
**[[2ri9]] - PcαMAN I + substrate analog<br /> | **[[2ri9]] - PcαMAN I + substrate analog<br /> | ||
**[[1krf]] - PcαMAN I + kifunensine + mannose<br /> | **[[1krf]] - PcαMAN I + kifunensine + mannose<br /> | ||
| - | + | ||
| - | + | ||
| - | + | ||
*α-MAN class II | *α-MAN class II | ||
| Line 76: | Line 83: | ||
*β-MAN | *β-MAN | ||
| + | **[[6dtt]] - mMAN <br /> | ||
| + | **[[6ddu]] - mMAN + β-mannose <br /> | ||
**[[2je8]] - BtβMAN residues 26-864 <br /> | **[[2je8]] - BtβMAN residues 26-864 <br /> | ||
**[[2wbk]] - BtβMAN residues 26-864 + mannopyranoside <br /> | **[[2wbk]] - BtβMAN residues 26-864 + mannopyranoside <br /> | ||
| Line 89: | Line 98: | ||
**[[5n6u]] - βMAN + β-mannose – ''Dictyoglomus thermophilum''<br /> | **[[5n6u]] - βMAN + β-mannose – ''Dictyoglomus thermophilum''<br /> | ||
**[[6byc]] - XaMAN – ''Xanthomonas axonopodis''<br /> | **[[6byc]] - XaMAN – ''Xanthomonas axonopodis''<br /> | ||
| - | **[[6byg]], [[ | + | **[[6byg]], [[6byi]] - XaMAN (mutant)<br /> |
**[[6bye]] - XaMAN + β-mannose<br /> | **[[6bye]] - XaMAN + β-mannose<br /> | ||
**[[4zxo]] - MAN – ''Bacterioides ovatus''<br /> | **[[4zxo]] - MAN – ''Bacterioides ovatus''<br /> | ||
| Line 121: | Line 130: | ||
**[[4nrr]] - RmβMAN (mutant) + mannosyl-fructose<br /> | **[[4nrr]] - RmβMAN (mutant) + mannosyl-fructose<br /> | ||
**[[4nrs]] - RmβMAN (mutant) + mannobiose<br /> | **[[4nrs]] - RmβMAN (mutant) + mannobiose<br /> | ||
| + | **[[6gvb]] - βMAN – ''Cutibacterium acnes''<br /> | ||
*Exo-α-1,6-MAN | *Exo-α-1,6-MAN | ||
| - | **[[5m7i]] - CpαMAN (mutant) + mannobiose – Clostridium perfringenes<br /> | + | **[[5m7i]] - CpαMAN (mutant) + mannobiose – ''Clostridium perfringenes''<br /> |
| - | **[[ | + | **[[5m7y]] - CpαMAN (mutant) + mannotriose <br /> |
| + | **[[6rqk]] - CpMAN + mannoimidazole<br /> | ||
*β-MAN/β-glucosidase | *β-MAN/β-glucosidase | ||
| - | **[[4re2]], [[4re3]], [[4re4]] - rβMAN/βGLU | + | **[[4re2]], [[4re3]], [[4re4]] - rβMAN/βGLU <br /> |
}} | }} | ||
Revision as of 09:55, 28 October 2019
| |||||||||||
3D structures of mannosidase
Updated on 28-October-2019
References
- ↑ Heikinheimo P, Helland R, Leiros HK, Leiros I, Karlsen S, Evjen G, Ravelli R, Schoehn G, Ruigrok R, Tollersrud OK, McSweeney S, Hough E. The structure of bovine lysosomal alpha-mannosidase suggests a novel mechanism for low-pH activation. J Mol Biol. 2003 Mar 28;327(3):631-44. PMID:12634058
- ↑ Ademark P, Lundqvist J, Hagglund P, Tenkanen M, Torto N, Tjerneld F, Stalbrand H. Hydrolytic properties of a beta-mannosidase purified from Aspergillus niger. J Biotechnol. 1999 Oct 8;75(2-3):281-9. PMID:10553664
- ↑ Berg T, Riise HM, Hansen GM, Malm D, Tranebjaerg L, Tollersrud OK, Nilssen O. Spectrum of mutations in alpha-mannosidosis. Am J Hum Genet. 1999 Jan;64(1):77-88. PMID:9915946 doi:http://dx.doi.org/S0002-9297(07)61660-7
- ↑ Uchino Y, Fukushige T, Yotsumoto S, Hashiguchi T, Taguchi H, Suzuki N, Konohana I, Kanzaki T. Morphological and biochemical studies of human beta-mannosidosis: identification of a novel beta-mannosidase gene mutation. Br J Dermatol. 2003 Jul;149(1):23-9. PMID:12890191
- ↑ Tankrathok A, Iglesias-Fernandez J, Luang S, Robinson RC, Kimura A, Rovira C, Hrmova M, Ketudat Cairns JR. Structural analysis and insights into the glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase. Acta Crystallogr D Biol Crystallogr. 2013 Oct;69(Pt 10):2124-35. doi:, 10.1107/S0907444913020568. Epub 2013 Sep 20. PMID:24100330 doi:http://dx.doi.org/10.1107/S0907444913020568