Tryptophan hydroxylase 1 with bound tryptophan
From Proteopedia
(Difference between revisions)
(New page: <StructureSection load="3e2t" size="400" color="" frame="true" spin="on" Scene='3e2t/3e2t_overall/2' align="right" caption= > 300px <!-- The line below this pa...) |
|||
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load="3e2t" size="400" color="" frame="true" spin="on" Scene='3e2t/3e2t_overall/2' | + | <StructureSection load="3e2t" size="400" color="" frame="true" spin="on" Scene='3e2t/3e2t_overall/2' side="right" caption= > |
[[Image:3e2t_1.jpg|left|300px]] | [[Image:3e2t_1.jpg|left|300px]] | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
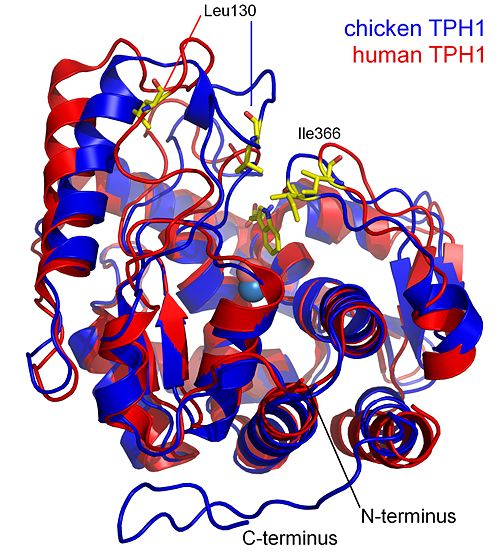
===The catalytic domain of chicken tryptophan hydroxylase 1 with bound tryptophan=== | ===The catalytic domain of chicken tryptophan hydroxylase 1 with bound tryptophan=== | ||
| - | [[Tryptophan hydroxylase]] is an iron and tetrahydrobiopterin dependent monooxygenase which belongs to the enzyme family of | + | [[Tryptophan hydroxylase]] is an iron and tetrahydrobiopterin dependent monooxygenase which belongs to the enzyme family of aromatic amino acid hydroxylases. |
The structure presented here is of the catalytic domain of chicken tryptophan hydroxylase 1 with bound tryptophan substrate. | The structure presented here is of the catalytic domain of chicken tryptophan hydroxylase 1 with bound tryptophan substrate. | ||
| Line 37: | Line 30: | ||
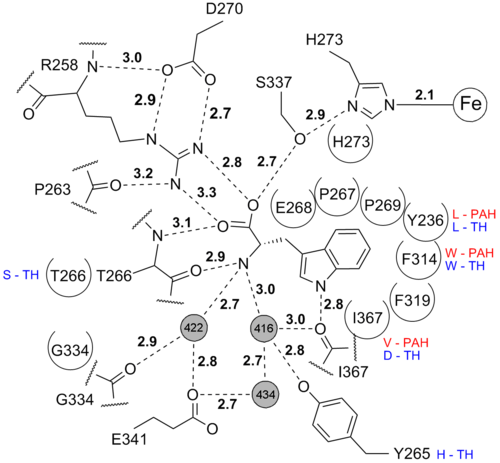
== Iron coordination == | == Iron coordination == | ||
| - | + | ||
The <scene name='User:Michael_Skovbo_Windahl/sandbox/3e2t_iron_coordination/3'>iron is coordinated</scene> by 2 histidines, one glutamate and one imidazole from the solvent. | The <scene name='User:Michael_Skovbo_Windahl/sandbox/3e2t_iron_coordination/3'>iron is coordinated</scene> by 2 histidines, one glutamate and one imidazole from the solvent. | ||
This coordination is called the 2-histidine-1-glutamate facial triad iron coordination and is seen in many mononuclear non-heme iron(II)enzymes<ref>PMID: 15739104</ref>. In this structure Glu317 coordinates the iron in a partial bidentate manner. The more common <scene name='User:Michael_Skovbo_Windahl/sandbox/1mlw_iron_coordination/3'>octahedral iron coordination</scene> (the resting state) for the 2-His-1-Glu iron coordination is seen in the structure of human TPH1 ([[1mlw]]). | This coordination is called the 2-histidine-1-glutamate facial triad iron coordination and is seen in many mononuclear non-heme iron(II)enzymes<ref>PMID: 15739104</ref>. In this structure Glu317 coordinates the iron in a partial bidentate manner. The more common <scene name='User:Michael_Skovbo_Windahl/sandbox/1mlw_iron_coordination/3'>octahedral iron coordination</scene> (the resting state) for the 2-His-1-Glu iron coordination is seen in the structure of human TPH1 ([[1mlw]]). | ||
| Line 47: | Line 40: | ||
<br/> | <br/> | ||
| - | <!-- | ||
| - | The line below this paragraph, {{ABSTRACT_PUBMED_18937498}}, adds the Publication Abstract to the page | ||
| - | (as it appears on PubMed at http://www.pubmed.gov), where 18937498 is the PubMed ID number. | ||
| - | --> | ||
{{ABSTRACT_PUBMED_18937498}} | {{ABSTRACT_PUBMED_18937498}} | ||
| - | + | </StructureSection> | |
==About this Structure== | ==About this Structure== | ||
3E2T is a 1 chain structure of sequence from [http://en.wikipedia.org/wiki/Gallus_gallus Gallus gallus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3E2T OCA]. | 3E2T is a 1 chain structure of sequence from [http://en.wikipedia.org/wiki/Gallus_gallus Gallus gallus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3E2T OCA]. | ||
| Line 78: | Line 67: | ||
[[Category: Petersen, C R.]] | [[Category: Petersen, C R.]] | ||
[[Category: Windahl, M S.]] | [[Category: Windahl, M S.]] | ||
| + | <br /> | ||
| + | Created with the participation of [[User:Michael Skovbo Windahl|Michael Skovbo Windahl]], [[User:Eran Hodis|Eran Hodis]], [[User:David Canner|David Canner]]. | ||
Current revision
| |||||||||||
About this Structure
3E2T is a 1 chain structure of sequence from Gallus gallus. Full crystallographic information is available from OCA.
Additional Resources
For additional information, see: Amino Acid Synthesis & Metabolism
References
- Windahl MS, Petersen CR, Christensen HE, Harris P. Crystal Structure of Tryptophan Hydroxylase with Bound Amino Acid Substrate. Biochemistry. 2008 Oct 21. PMID:18937498 doi:10.1021/bi8015263
- Nielsen MS, Petersen CR, Munch A, Vendelboe TV, Boesen J, Harris P, Christensen HE. A simple two step procedure for purification of the catalytic domain of chicken tryptophan hydroxylase 1 in a form suitable for crystallization. Protein Expr Purif. 2008 Feb;57(2):116-26. Epub 2007 Nov 20. PMID:18055219 doi:10.1016/j.pep.2007.10.016
- ↑ Jiang GC, Yohrling GJ 4th, Schmitt JD, Vrana KE. Identification of substrate orienting and phosphorylation sites within tryptophan hydroxylase using homology-based molecular modeling. J Mol Biol. 2000 Sep 29;302(4):1005-17. PMID:10993738 doi:10.1006/jmbi.2000.4097
- ↑ McKinney J, Teigen K, Froystein NA, Salaun C, Knappskog PM, Haavik J, Martinez A. Conformation of the substrate and pterin cofactor bound to human tryptophan hydroxylase. Important role of Phe313 in substrate specificity. Biochemistry. 2001 Dec 25;40(51):15591-601. PMID:11747434
- ↑ Daubner SC, Moran GR, Fitzpatrick PF. Role of tryptophan hydroxylase phe313 in determining substrate specificity. Biochem Biophys Res Commun. 2002 Apr 5;292(3):639-41. PMID:11922614 doi:10.1006/bbrc.2002.6719
- ↑ Koehntop KD, Emerson JP, Que L Jr. The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J Biol Inorg Chem. 2005 Mar;10(2):87-93. Epub 2005 Mar 1. PMID:15739104 doi:10.1007/s00775-005-0624-x
Created with the participation of Michael Skovbo Windahl, Eran Hodis, David Canner.