UDP-N-acetylglucosamine acyltransferase
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
**[[5f42]] – LpxA – ''Francicella novicida''<br /> | **[[5f42]] – LpxA – ''Francicella novicida''<br /> | ||
| - | **[[5dem]] – PaLpxA – ''Pseudomonas aeruginosa''<br /> | + | **[[5dem]], [[6ued]] – PaLpxA – ''Pseudomonas aeruginosa''<br /> |
**[[4r36]] – BfLpxA – ''Bacterioides fragilis''<br /> | **[[4r36]] – BfLpxA – ''Bacterioides fragilis''<br /> | ||
**[[4e6t]], [[4e6u]] – LpxA – ''Acinetobacter baumannii''<br /> | **[[4e6t]], [[4e6u]] – LpxA – ''Acinetobacter baumannii''<br /> | ||
| Line 26: | Line 26: | ||
**[[1lxa]] – EcLpxA (mutant)<br /> | **[[1lxa]] – EcLpxA (mutant)<br /> | ||
**[[1j2z]] – LpxA – ''Helicobacter pylori''<br /> | **[[1j2z]] – LpxA – ''Helicobacter pylori''<br /> | ||
| + | **[[5jxx]] – LpxA – ''Moraxella catarrhalis''<br /> | ||
| + | **[[6oss]] – LpxA – ''Proteus Mirabilis''<br /> | ||
**[[5jxx]] – LpxA – ''Moraxella catarrhalis''<br /> | **[[5jxx]] – LpxA – ''Moraxella catarrhalis''<br /> | ||
| Line 32: | Line 34: | ||
**[[5dep]], [[5dg3]] – PaLpxA + UDP-GlcNAc derivative<br /> | **[[5dep]], [[5dg3]] – PaLpxA + UDP-GlcNAc derivative<br /> | ||
**[[4r37]] – BfLpxA + UDP-GlcNAc <br /> | **[[4r37]] – BfLpxA + UDP-GlcNAc <br /> | ||
| + | **[[6uec]], [[6uee]], [[6ueg]] – PaLpxA + ligand<br /> | ||
**[[4j09]] – EcLpxA + RJPXD33 peptide <br /> | **[[4j09]] – EcLpxA + RJPXD33 peptide <br /> | ||
| - | **[[ | + | **[[6p9p]] – EcLpxA + inhibitor<br /> |
**[[2jf3]] – EcLpxA + UDP-GlcNAc <br /> | **[[2jf3]] – EcLpxA + UDP-GlcNAc <br /> | ||
**[[3i3a]], [[3i3x]] – LiLpxA + sugar nucleotide<br /> | **[[3i3a]], [[3i3x]] – LiLpxA + sugar nucleotide<br /> | ||
| - | ** | + | **[[2qiv]], [[2jf3]] – EcLpxA + sugar nucleotide<br /> |
| + | **[[6p9q]], [[8p9r]], [[6p9s]], [[6p9t]] - EcLpxA + inhibitor + UDP-myristoyl-GlcNAc<br /> | ||
| + | |||
| + | |||
}} | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 11:26, 19 March 2020
| |||||||||||
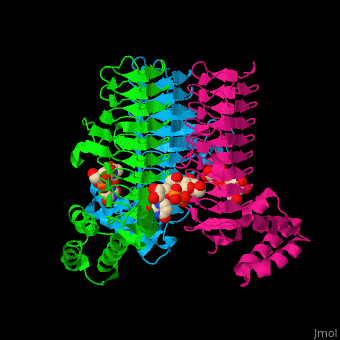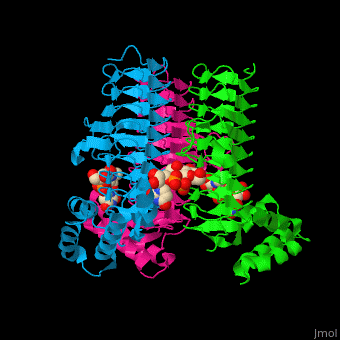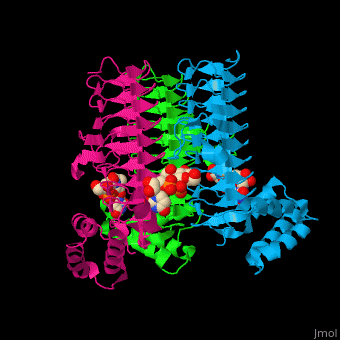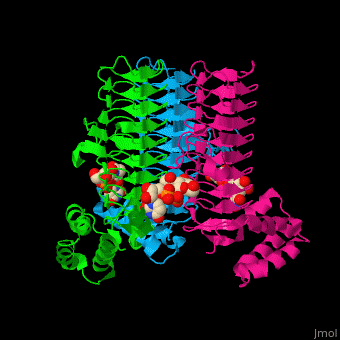
3D structures of UDP-N-acetylglucosamine acyltransferase
Updated on 19-March-2020
References
- ↑ Wyckoff TJ, Raetz CR. The active site of Escherichia coli UDP-N-acetylglucosamine acyltransferase. Chemical modification and site-directed mutagenesis. J Biol Chem. 1999 Sep 17;274(38):27047-55. PMID:10480918
- ↑ Ulaganathan V, Buetow L, Hunter WN. Nucleotide substrate recognition by UDP-N-acetylglucosamine acyltransferase (LpxA) in the first step of lipid A biosynthesis. J Mol Biol. 2007 Jun 1;369(2):305-12. Epub 2007 Mar 21. PMID:17434525 doi:10.1016/j.jmb.2007.03.039