|
|
| (19 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | <StructureSection size="450" scene ="Journal:JBIC:5/Opening/2" caption="Solved Crystal Structure of Hyperactive Catechol Dioxygenase"> | + | <StructureSection size="" scene ="Journal:JBIC:5/Opening/2" caption="Solved Crystal Structure of Hyperactive Catechol Dioxygenase"> |
| | + | |
| | + | __TOC__ |
| | + | |
| | + | == Function == |
| | | | |
| | '''Dioxygenases''' cleave the aromatic rings of their substrates by inserting two oxygen atoms, thus degrading these compounds. The dioxygenases are divided into 2 groups according to their mode of ring scission. The intradiol enzymes use Fe+3 as cofactor and cleave the substrate between 2 hydroxyl groups. The extradiol enzymes use Fe+2 as cofactor and cleave the substrate between a hydroxylated carbon and a non-hydroxylated one. <br /> | | '''Dioxygenases''' cleave the aromatic rings of their substrates by inserting two oxygen atoms, thus degrading these compounds. The dioxygenases are divided into 2 groups according to their mode of ring scission. The intradiol enzymes use Fe+3 as cofactor and cleave the substrate between 2 hydroxyl groups. The extradiol enzymes use Fe+2 as cofactor and cleave the substrate between a hydroxylated carbon and a non-hydroxylated one. <br /> |
| Line 7: |
Line 11: |
| | * '''Naphthalene 1,2-dioxygenase''' (NDO) catalyzes the conversion of naphthalene and O2 to dihydronaphthalene-diol.<br /> | | * '''Naphthalene 1,2-dioxygenase''' (NDO) catalyzes the conversion of naphthalene and O2 to dihydronaphthalene-diol.<br /> |
| | * '''Glyoxalase/bleomycin resistant protein/dioxygenase''' (GBD) catalyzes the cleavage of S-lactoyl-glutathione to methylglyoxal and glutathione.<br /> | | * '''Glyoxalase/bleomycin resistant protein/dioxygenase''' (GBD) catalyzes the cleavage of S-lactoyl-glutathione to methylglyoxal and glutathione.<br /> |
| - | * '''α-ketoglutarate-dependent dioxygenase homolog 5''' (Alkbh5) is an RNA demethylase which reverses the N(6)-methyladenosine modification of RNA. | + | * '''Methylcytosine dioxygenase''' (TET) catalyzes the conversion of 5-methylcytosine to 5-hydroxymethylcytosine to mediate DNA demethylation<ref>PMID:25862091</ref>. For details see [[TET Enzymes]].<br /> |
| | + | * '''α-ketoglutarate-dependent dioxygenase Alkb''' repairs alkylated DNA and RNA containing 3-methylcytosine or 1-methyladenine by oxidative demethylation<ref>PMID:18432238</ref>.<br /> |
| | + | * '''α-ketoglutarate-dependent dioxygenase Fto''' highest activity is repairing alkylated RNA containing 3-methyluracil by oxidative demethylation<ref>PMID:18775698</ref>.<br /> |
| | | | |
| - | === A Hyperactive Cobalt-Substituted Extradiol-Cleaving Catechol Dioxygenase <ref>DOI 10.1007/s00775-010-0732-0</ref>===
| + | == A Hyperactive Cobalt-Substituted Extradiol-Cleaving Catechol Dioxygenase <ref>DOI 10.1007/s00775-010-0732-0</ref>== |
| | | | |
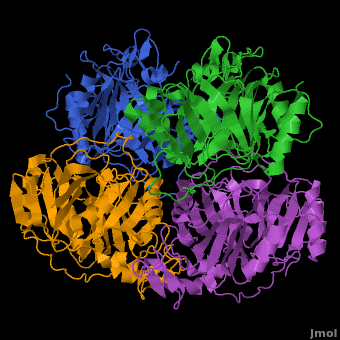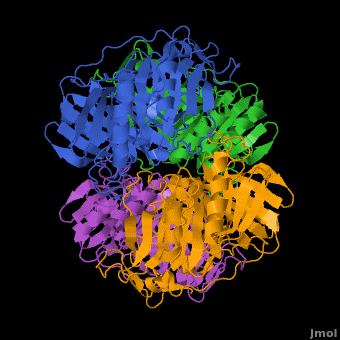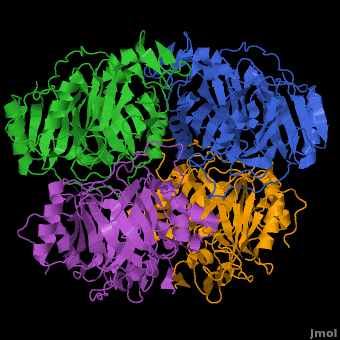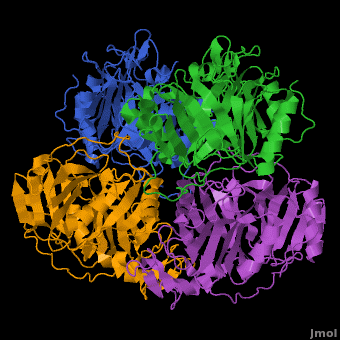
| | In most metalloenzyme mechanisms, O<sub>2</sub> is reductively activated by direct electron transfer from the reduced metal ion. This implies that the rate of the reaction should be strongly dependent on the redox potential of the catalytic metal. We have proposed a different strategy for the Fe(II)-containing extradiol aromatic ring-cleaving dioxygenase family in which the activating electron is derived from the aromatic substrate rather the iron. When the aromatic substrate and the O<sub>2</sub> are bound to the metal at the same time, the metal may serve as a conduit for the activating electron. Since there is no net change in metal oxidation state during this process, the impact of the redox potential of the metal on oxygen activation should be minimal, so long as it is low enough to allow formation of the initial O<sub>2</sub> complex. This hypothesis is tested here by substituting Co(II) for Fe(II) in the <scene name='Journal:JBIC:5/Cobalt_overview/1'>Homoprotocatechuate 2,3-dioxygenase</scene> (2,3-HPCD) from ''Brevibacterium fuscum''. | | In most metalloenzyme mechanisms, O<sub>2</sub> is reductively activated by direct electron transfer from the reduced metal ion. This implies that the rate of the reaction should be strongly dependent on the redox potential of the catalytic metal. We have proposed a different strategy for the Fe(II)-containing extradiol aromatic ring-cleaving dioxygenase family in which the activating electron is derived from the aromatic substrate rather the iron. When the aromatic substrate and the O<sub>2</sub> are bound to the metal at the same time, the metal may serve as a conduit for the activating electron. Since there is no net change in metal oxidation state during this process, the impact of the redox potential of the metal on oxygen activation should be minimal, so long as it is low enough to allow formation of the initial O<sub>2</sub> complex. This hypothesis is tested here by substituting Co(II) for Fe(II) in the <scene name='Journal:JBIC:5/Cobalt_overview/1'>Homoprotocatechuate 2,3-dioxygenase</scene> (2,3-HPCD) from ''Brevibacterium fuscum''. |
| | | | |
| | 2,3-HPCD catalyzes the cleavage of the C2-C3 bond of homoprotocatechuate (HPCA) with the incorporation of both oxygen atoms derived from O<sub>2</sub> to form ring opened, 5-carboxymethyl-2-hydroxymuconic semialdehyde (5-CHMSA). Despite the significantly higher standard redox potential of Co(III/II) (1.92 V versus SHE) relative to that of the native metal Fe(II) (0.77 V), Co-HPCD was found to have more than twice the specific activity of Fe-HPCD under saturating O<sub>2</sub> conditions, making Co-HPCD a hyper-active enzyme. The presence of <scene name='Journal:JBIC:5/Cobalt_4_sites/3'>Co in the active sites of Co-HPCD</scene> is illustrated by the 4 strong anomalous difference peaks determined from the X-ray diffraction data set collected at the cobalt K-edge (Figure 4A, 1.6050 Å, 7.725 keV), which shows cobalt coordinated via <scene name='Journal:JBIC:5/Hpcd_active_site/5'>2-His-1-Carboxylate facial triad</scene>. The structural comparison of the high-resolution X-ray crystal structures of Fe-HPCD (1.70 Å, [[3ojt]]), Mn-HPCD (1.65 Å, [[3ojn]]), and Co-HCPD (1.72 Å, [[3ojj]]) shows <scene name='Journal:JBIC:5/Overlays/16'>no significant structural differences in the active site</scene> environment as indicated by the RMSD values of 0.10-0.14 Å for superposition of <scene name='Journal:JBIC:5/Overlays_15a/10'>all atoms within 15 angstroms of the metal center</scene>. In addition, the presence of Co in the active site of HPCD has <scene name='Journal:JBIC:5/Substrate_intro/6'>no structural consequences</scene> on either the <scene name='Journal:JBIC:5/Substrate_overlay/15'>mode of substrate binding</scene> or on the conformational integrity of the active site residues in<scene name='Journal:JBIC:5/Substrate_overlay/16'> the 1st or 2nd coordination sphere environment</scene>. The absence of any observable structural differences upon metal substitutions suggests that differential redox tuning of the metal centers in this dioxygenase is highly unlikely. Rather, the 2,3-HPCD enzyme can carry out the O<sub>2</sub> activation and oxidative ring-cleavage efficiently over a large range of metal redox potentials. The structural analysis supports the proposed mechanism described above in which the ability of the enzyme to activate molecular O<sub>2</sub> does not correlate with redox potential of the metal center. However, the current results also show that the rates of individual steps in the overall catalytic cycle can be effected by the metal present in the active site of HPCD, resulting in a earlier rate-limiting step for Co-HPCD compared to Fe-HPCD and Mn-HPCD. | | 2,3-HPCD catalyzes the cleavage of the C2-C3 bond of homoprotocatechuate (HPCA) with the incorporation of both oxygen atoms derived from O<sub>2</sub> to form ring opened, 5-carboxymethyl-2-hydroxymuconic semialdehyde (5-CHMSA). Despite the significantly higher standard redox potential of Co(III/II) (1.92 V versus SHE) relative to that of the native metal Fe(II) (0.77 V), Co-HPCD was found to have more than twice the specific activity of Fe-HPCD under saturating O<sub>2</sub> conditions, making Co-HPCD a hyper-active enzyme. The presence of <scene name='Journal:JBIC:5/Cobalt_4_sites/3'>Co in the active sites of Co-HPCD</scene> is illustrated by the 4 strong anomalous difference peaks determined from the X-ray diffraction data set collected at the cobalt K-edge (Figure 4A, 1.6050 Å, 7.725 keV), which shows cobalt coordinated via <scene name='Journal:JBIC:5/Hpcd_active_site/5'>2-His-1-Carboxylate facial triad</scene>. The structural comparison of the high-resolution X-ray crystal structures of Fe-HPCD (1.70 Å, [[3ojt]]), Mn-HPCD (1.65 Å, [[3ojn]]), and Co-HCPD (1.72 Å, [[3ojj]]) shows <scene name='Journal:JBIC:5/Overlays/16'>no significant structural differences in the active site</scene> environment as indicated by the RMSD values of 0.10-0.14 Å for superposition of <scene name='Journal:JBIC:5/Overlays_15a/10'>all atoms within 15 angstroms of the metal center</scene>. In addition, the presence of Co in the active site of HPCD has <scene name='Journal:JBIC:5/Substrate_intro/6'>no structural consequences</scene> on either the <scene name='Journal:JBIC:5/Substrate_overlay/15'>mode of substrate binding</scene> or on the conformational integrity of the active site residues in<scene name='Journal:JBIC:5/Substrate_overlay/16'> the 1st or 2nd coordination sphere environment</scene>. The absence of any observable structural differences upon metal substitutions suggests that differential redox tuning of the metal centers in this dioxygenase is highly unlikely. Rather, the 2,3-HPCD enzyme can carry out the O<sub>2</sub> activation and oxidative ring-cleavage efficiently over a large range of metal redox potentials. The structural analysis supports the proposed mechanism described above in which the ability of the enzyme to activate molecular O<sub>2</sub> does not correlate with redox potential of the metal center. However, the current results also show that the rates of individual steps in the overall catalytic cycle can be effected by the metal present in the active site of HPCD, resulting in a earlier rate-limiting step for Co-HPCD compared to Fe-HPCD and Mn-HPCD. |
| - | </StructureSection> | |
| | | | |
| - | ==3D structures of Protocatechuate 3,4-dioxygenase== | + | ==3D structures of dioxygenase== |
| | + | [[Dioxygenase 3D structures]] |
| | | | |
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}}
| + | </StructureSection> |
| - | {{#tree:id=OrganizedByTopic|openlevels=0|
| + | ==References== |
| - | | + | |
| - | *'''Dioxygenase Intradiol family'''
| + | |
| - | | + | |
| - | *Protocatechuate 3,4-dioxygenase (PCD)
| + | |
| - | | + | |
| - | **[[2pcd]], [[4who]], [[4whp]] – PpPCD α,β + Fe+3 – ''Pseudomonas putida''<br />
| + | |
| - | **[[3pcd]], [[1ykk]], [[1ykm]], [[1yko]], [[3lkt]], [[3mfl]], [[3mi1]], [[3mv4]], [[3t63]] – PpPCD α,β (mutant) + Fe+3 <br />
| + | |
| - | **[[1eo2]], [[1eo9]], [[1eoa]], [[2bum]], [[2but]] – AcPCD α,β + Fe+3 - ''Acinetobacter''<br />
| + | |
| - | **[[2bux]] – AcPCD α,β (mutant) + Fe+3 <br />
| + | |
| - | | + | |
| - | *''PCD complex with aromatic compound''
| + | |
| - | | + | |
| - | **[[3pcf]], [[3pch]], [[3pci]], [[4whq]], [[4whr]], [[4whs]] – PpPCD α,β + Fe+3 + HB derivative<br />
| + | |
| - | **[[3pca]], [[3pcb]], [[3pcc]] – PpPCD α,β + Fe+3 + HB <br />
| + | |
| - | **[[1ykl]], [[1ykn]], [[1ykp]], [[3lmx]], [[3mv6]] – PpPCD α,β (mutant) + Fe+3 + DHB<br />
| + | |
| - | **[[3pcj]], [[3pck]], [[3pcl]], [[3pcm]] – PpPCD α,β + Fe+3 + hydroxynicotinic acid derivative<br />
| + | |
| - | **[[3pcn]], [[3pce]], [[3pcg]] – PpPCD α,β + Fe+3 + hydroxyphenyl acetate derivative<br />
| + | |
| - | **[[1eob]] – AcPCD α,β + Fe+3 + DHB <br />
| + | |
| - | **[[2buv]], [[2bv0]] – AcPCD α,β (mutant) + Fe+3 + DHB <br />
| + | |
| - | **[[2bur]] – AcPCD α,β + Fe+3 + HB <br />
| + | |
| - | **[[2buw]] – AcPCD α,β (mutant) + Fe+3 + HB <br />
| + | |
| - | **[[1eoc]], [[2buq]] – AcPCD α,β + Fe+3 + catechol derivative<br />
| + | |
| - | **[[2buu]], [[2buy]], [[2buz]] – AcPCD α,β (mutant) + Fe+3 + catechol derivative<br />
| + | |
| - | **[[3lxv]], [[3mi5]], [[3t67]] – PpPCD α,β (mutant) + Fe+3 + catechol derivative<br />
| + | |
| - | | + | |
| - | *Protocatechuate 4,5-dioxygenase (PCD)
| + | |
| - | | + | |
| - | **[[1b4u]] – SpPCD α,β + Fe+3 + DHB – ''Sphingomonas paucimobilis''<br />
| + | |
| - | **[[1bou]] – SpPCD α,β + Fe+3 <br />
| + | |
| - | | + | |
| - | *Catechol 1,2-dioxygenase (CTD)
| + | |
| - | | + | |
| - | **[[1dlm]], [[1dlq]] – AcCTD + Fe+3 <br />
| + | |
| - | **[[2azq]] – PpCTD + Fe+3 <br />
| + | |
| - | | + | |
| - | *''CTD complex with aromatic compound''
| + | |
| - | | + | |
| - | **[[1dlt]], [[1dmh]] – AcCTD + Fe+3 + catechol derivative<br />
| + | |
| - | **[[3hgi]] – RoCTD + Fe+3 + benzoic acid <br />
| + | |
| - | **[[3hhy]], [[3hkp]] – RoCTD + Fe+3 + DHB<br />
| + | |
| - | **[[3hj8]], [[3hjq]], [[3hjs]], [[3i4v]], [[3i4y]], [[3i51]] – RoCTD + Fe+3 + catechol derivative<br />
| + | |
| - | **[[3hhx]] – RoCTD + Fe+3 + pyrogallol<br />
| + | |
| - | **[[2xsu]] – ArCTD + Fe+3 + phosphoinositol derivative – ''Acinetobacter radioresistens''<br />
| + | |
| - | **[[2xsv]], [[2xsr]] – ArCTD + Fe+3 + phosphoinositol derivative <br />
| + | |
| - | | + | |
| - | *Chlorocatechol 1,2-dioxygenase (CCTD)
| + | |
| - | | + | |
| - | **[[3th1]] – PpCCTD + Fe+3 <br />
| + | |
| - | **[[1s9a]] – RoCCTD + Fe+3 <br />
| + | |
| - | **[[3o32]], [[3o5u]] – RoCCTD + Fe+3 + dichlorocatechol derivative<br />
| + | |
| - | **[[3o6j]] – RoCCTD + Fe+3 + hydroxyquinol<br />
| + | |
| - | **[[3o6r]] – RoCCTD + Fe+3 + pyrogallol<br />
| + | |
| - | **[[2boy]] – RoCCTD + Fe+3 + benzhydroxamate<br />
| + | |
| - | | + | |
| - | *Catechol 2,3-dioxygenase (CTD)
| + | |
| - | | + | |
| - | **[[2wl3]], [[2wl9]] - RoCTD + Fe+3 - ''Rhodococcus opacus'' <br />
| + | |
| - | **[[3lm4]] – RoCTD + Fe+3 + H2O2<br />
| + | |
| - | | + | |
| - | *Hydroxyquinol 1,2-dioxygenase (HQD)
| + | |
| - | | + | |
| - | **[[1tmx]] – PpHQD + Fe+3 – ''Pimelobacter simplex''<br />
| + | |
| - | | + | |
| - | *Nitrobenzene dioxygenase (NBDO)
| + | |
| - | | + | |
| - | **[[2bmo]] – CtNBDO + Fe+3 + Fe2S2 <br />
| + | |
| - | **[[2bmq]] – CtNBDO + Fe+3 + Fe2S2 + nitrobenzene<br />
| + | |
| - | **[[2bmr]] – CtNBDO + Fe+3 + Fe2S2 + nitrotoluene<br />
| + | |
| - | | + | |
| - | *α-ketoglutarate-dependent dioxygenase (Alkb)
| + | |
| - | | + | |
| - | **[[3t4h]], [[3t4v]], [[3t3y]] – EcAlkb + Fe+3 + inhibitor – ''Escherichia coli''<br />
| + | |
| - | **[[2fdj]], [[2fdg]] – EcAlkb + Mn+2 + succinate <br />
| + | |
| - | **[[4jht]] – EcAlkb + Mn+2 + qinoline derivative <br />
| + | |
| - | **[[2fdf]] – EcAlkb (mutant) + Co+2 + oxoglutarate <br />
| + | |
| - | **[[2fd8]] – EcAlkb (mutant) + Fe+2 + oxoglutarate <br />
| + | |
| - | **[[3khb]] – hAlkb (mutant) + Co+2 + oxoglutarate <br />
| + | |
| - | **[[3i3q]] – hAlkb + Mn+2 + oxoglutarate <br />
| + | |
| - | **[[3thp]] – hAlkb + Mn+2 + Zn+2 + oxoglutarate <br />
| + | |
| - | **[[3tht]] – hAlkb (mutant) + Mn+2 + Zn+2 + oxoglutarate <br />
| + | |
| - | **[[4idz]] – hFto + Ni+2 + oxalylglycine<br />
| + | |
| - | **[[4ie0]], [[4ie5]] – hFto + Zn+2 + pyrimidine derivative<br />
| + | |
| - | **[[4ie4]], [[4ie6]] – hFto + Zn+2 + quinoline derivative<br />
| + | |
| - | **[[4ie7]] – hFto + Zn+2 + rhein<br />
| + | |
| - | **[[4qkn]] – hFto + Mn+2 + meclofenamic acid<br />
| + | |
| - | **[[4cxw]], [[4cxx]], [[4cxy]] – hFto + Ni+2 + inhibitor<br />
| + | |
| - | | + | |
| - | *''Alkb complex with DNA''
| + | |
| - | | + | |
| - | **[[3rzm]], [[3rzl]], [[3rzh]], [[3rzg]], [[3btz]], [[4mg2]], [[4mgt]] – hAlkb (mutant) + DNA<br />
| + | |
| - | **[[3h8x]], [[3h8r]], [[3h8o]], [[3bty]] – hAlkb (mutant) + DNA + inhibitor<br />
| + | |
| - | **[[3btx]] – hAlkb (mutant) + Mg+2 + DNA <br />
| + | |
| - | **[[3i49]], [[3i2o]] – hAlkb + Fe+2 + oxoglutarate + DNA <br />
| + | |
| - | **[[4nid]] – hAlkb + Mn+2 + oxoglutarate + DNA <br />
| + | |
| - | **[[4nig]], [[4nih]], [[4nii]] – hAlkb (mutant) + Mn+2 + oxoglutarate + DNA <br />
| + | |
| - | **[[3o1s]] – hAlkb + DNA <br />
| + | |
| - | **[[3i3m]] – hAlkb + Mn+2 + oxoglutarate + DNA<br />
| + | |
| - | **[[3s5a]], [[3s57]], [[3rzk]], [[3rzj]], [[3buc]], [[3bu0]], [[3bkz]] – hAlkb (mutant) + Mn+2 + oxoglutarate + DNA<br />
| + | |
| - | **[[3o1r]], [[3o1p]], [[3o1o]], [[3o1m]] – hAlkb (mutant) + Mn+2 + oxoglutarate + DNA + inhibitor<br />
| + | |
| - | **[[3bie]], [[3bi3]], [[2fdh]] – EcAlkb (mutant) + Mn+2 + oxoglutarate + DNA + inhibitor<br />
| + | |
| - | **[[2fdi]] – EcAlkb + Fe+2 + oxoglutarate + DNA + inhibitor<br />
| + | |
| - | **[[3khc]] – hAlkb (mutant) + Co+2 + oxoglutarate + DNA + inhibitor<br />
| + | |
| - | **[[3o1v]], [[3o1u]], [[3o1t]], [[3t1s]] – hAlkb (mutant) + Fe+3 + succinate + DNA + inhibitor<br />
| + | |
| - | | + | |
| - | *α-ketoglutarate-dependent dioxygenase homolog 5 (Alkbh5)
| + | |
| - | | + | |
| - | **[[4nj4]], [[4o7x]] – hAlkbh5 residues 66-292 + Mn<br />
| + | |
| - | **[[4o61]] – hAlkbh5 residues 74-294 + citrate<br />
| + | |
| - | **[[4nrm]] – hAlkbh5 residues 66-292 + citrate + acetate<br />
| + | |
| - | **[[4nro]] – hAlkbh5 residues 66-292 + Mn + ketoglutarate <br />
| + | |
| - | **[[4oct]] – hAlkbh5 residues 66-292 + Mn + oxoglutarate <br />
| + | |
| - | **[[4nrp]] – hAlkbh5 residues 66-292 + Mn + oxalylglycine <br />
| + | |
| - | **[[4nrq]] – hAlkbh5 residues 66-292 + Mn + pyridine dicarboxylate <br />
| + | |
| - | **[[4npl]] – zfAlkbh5 residues 38-287 + Mn + ketoglutarate – zebra fish<br />
| + | |
| - | **[[4npm]] – zfAlkbh5 residues 38-287 + Mn + succinate <br />
| + | |
| - | | + | |
| - | *Leucoanthocyanidin dioxygenase (LACD)
| + | |
| - | | + | |
| - | **[[1gp5]], [[1gp6]] – AtLACD + dihydroquercetin – ''Arabidopsis thaliana'' <br />
| + | |
| - | **[[2brt]] – AtLACD + naringenin <br />
| + | |
| - | | + | |
| - | *'''Dioxygenase extradiol family'''
| + | |
| - | | + | |
| - | *Homoprotocatechuate 2,3-dioxygenase (HPCD)
| + | |
| - | | + | |
| - | **[[1f1r]], [[1f1u]] – AgHPCD + Mn+2 – ''Arthrobacter globiformis''<br />
| + | |
| - | **[[1f1x]], [[1q0o]] – BfHPCD + Fe+3 – ''Brevibacterium fuscum''<br />
| + | |
| - | **[[2ig9]], [[3ojt]] – BfHPCD + Fe+2<br />
| + | |
| - | **[[3ecj]], [[4ghc]] – BfHPCD (mutant) + Fe+2<br />
| + | |
| - | **[[3bza]], [[3ojn]] – BfHPCD + Mn+2<br />
| + | |
| - | **[[3ojj]] – BfHPCD + MCo+2<br />
| + | |
| - | | + | |
| - | *''HPCD complex with aromatic compound''
| + | |
| - | | + | |
| - | **[[1f1v]] – AgHPCD + Mn+2 + dihydroxyphenyl acetate <br />
| + | |
| - | **[[1q0c]], [[4ghg]] – BfHPCD + Fe+2 + dihydroxyphenyl acetate <br />
| + | |
| - | **[[4ghd]] – BfHPCD (mutant) + Fe+2 + dihydroxyphenyl acetate <br />
| + | |
| - | **[[2iga]] – BfHPCD + Fe+2 + O2 + reaction intermediates<br />
| + | |
| - | **[[3eck]] – BfHPCD (mutant) + Fe+2 + reaction intermediates<br />
| + | |
| - | **[[3ojk]] – BfHPCD + Co+2 + catechol derivative<br />
| + | |
| - | **[[4ghe]], [[4ghf]] – BfHPCD (mutant) + Fe+2 + catechol derivative<br />
| + | |
| - | **[[4ghh]] – BfHPCD + Fe+2 + catechol derivative<br />
| + | |
| - | | + | |
| - | *Catechol 2,3-dioxygenase (MPC)
| + | |
| - | | + | |
| - | **[[1mpy]] – PpMPC + Fe+2 <br />
| + | |
| - | **[[3hpv]] – PsMPC + Fe+2 - ''Pseudomonas''<br />
| + | |
| - | **[[3hpy]] – PsMPC + Fe+2 + catechol derivative<br />
| + | |
| - | **[[3hq0]] – PsMPC + Fe+3 + product<br />
| + | |
| - | | + | |
| - | *Naphthalene 1,2-dioxygenase (NDO)
| + | |
| - | | + | |
| - | **[[1ndo]], [[1o7h]], [[1o7w]] – PpNDO + Fe+3 + Fe2S2<br />
| + | |
| - | **[[2hmj]] – PpNDO (mutant) + Fe+3 + Fe2S2 <br />
| + | |
| - | **[[2b1x]] – RoNDO + Fe+3 + Fe2S2<br />
| + | |
| - | | + | |
| - | *''NDO complex with aromatic compound''
| + | |
| - | | + | |
| - | **[[1eg9]] – PpNDO + Fe+3 + Fe2S2 + indole<br />
| + | |
| - | **[[1o7g]] – PpNDO + Fe+3 + Fe2S2 + naphthalene<br />
| + | |
| - | **[[1o7m]] – PpNDO + Fe+3 + Fe2S2 + O2<br />
| + | |
| - | **[[1o7n]] – PpNDO + Fe+3 + Fe2S2 + O2 + indole<br />
| + | |
| - | **[[1o7p]] – PpNDO + Fe+3 + Fe2S2 + dihydronaphthalene<br />
| + | |
| - | **[[1uuv]], [[1uuw]] – PpNDO + Fe+3 + Fe2S2 + NO3<br />
| + | |
| - | **[[2hmk]], [[2hml]] – PpNDO (mutant) + Fe+3 + Fe2S2 + phenanthrene<br />
| + | |
| - | **[[2hmm]], [[2hmn]] – PpNDO (mutant) + Fe+3 + Fe2S2 + anthracene<br />
| + | |
| - | **[[2hmo]] – PpNDO + Fe+3 + Fe2S2 + nitrotoluene<br />
| + | |
| - | **[[2b24]] – RoNDO + Fe+3 + Fe2S2 + indole<br />
| + | |
| - | **[[4hm0]], [[4hm1]], [[4hm4]], [[4hm5]] – PsNDO + Fe+3 + Fe2S2 + indole derivative<br />
| + | |
| - | **[[4hm2]], [[4hm3]], [[4hm6]], [[4hm7]], [[4hm8]] – PsNDO + Fe+3 + Fe2S2 + phenyl derivative<br />
| + | |
| - | **[[4hjl]] – PsNDO + Fe+3 + Fe2S2 + chloronaphthalene<br />
| + | |
| - | **[[4hkv]] – PsNDO + Fe+3 + Fe2S2 + benzamide<br />
| + | |
| - | | + | |
| - | *Dihydronaphthalene 1,2-dioxygenase (HNDO)
| + | |
| - | | + | |
| - | **[[2ehz]], [[2ei2]] – PsHNDO + Fe+2 <br />
| + | |
| - | **[[2ei0]], [[2ei1]], [[2ei3]] – PsHNDO + Fe+2 + dihydroxybephenyl<br />
| + | |
| - | | + | |
| - | *2,3-dihydroxybiphenyl 1,2-dioxygenase (DHBP)
| + | |
| - | | + | |
| - | **[[1dhy]], [[1eil]], [[1eiq]], [[1eir]] – PsDHBP + Fe+3 <br />
| + | |
| - | **[[1kw3]] – PsDHBP + Fe+2 <br />
| + | |
| - | **[[1kw8]] – PsDHBP + Fe+2 + NO<br />
| + | |
| - | **[[1kwb]] – PsDHBP (mutant) <br />
| + | |
| - | **[[1han]], [[1kmy]] – BxDHBP (mutant) + Fe+3 – ''Burkholderia xenovorans''<br />
| + | |
| - | | + | |
| - | *''DHBP complex with aromatic compound''
| + | |
| - | | + | |
| - | **[[1knd]], [[1knf]] – BxDHBP + Fe+3 + catechol derivative<br />
| + | |
| - | **[[1lgt]], [[1lkd]] – BxDHBP + Fe+2 + dihydroxybephenyl<br />
| + | |
| - | **[[1kw6]], [[1kw9]] – PsDHBP + Fe+2 + dihydroxybephenyl<br />
| + | |
| - | **[[1kwc]] – PsDHBP (mutant) + dihydroxybephenyl<br />
| + | |
| - | | + | |
| - | *Salicylate 1,2-dioxygenase (SDO)
| + | |
| - | | + | |
| - | **[[2phd]] – PssSDO + Fe+3 – ''Pseudaminobacter salicylatoxidans''<br />
| + | |
| - | **[[4fah]], [[4fbf]] – PssSDO (mutant) + Fe+3<br />
| + | |
| - | **[[3nst]] – PssSDO (mutant) + Fe+2 <br />
| + | |
| - | **[[3njz]] – PssSDO + Fe+2 +salicylate<br />
| + | |
| - | **[[3nvc]] – PssSDO (mutant) + Fe+2 +salicylate<br />
| + | |
| - | **[[3nkt ]] – PssSDO + Fe+2 +naphthoate<br />
| + | |
| - | **[[3nl1]] – PssSDO + Fe+2 +gentisate<br />
| + | |
| - | **[[3nw4]], [[4fag]] – PssSDO (mutant) + Fe+2 +gentisate<br />
| + | |
| - | | + | |
| - | *Biphenyl 1,2-dioxygenase (BDO)
| + | |
| - | | + | |
| - | **[[3gzy]] – CtBDO + Fe+2 + Fe2S2 – ''Comamonas testosteroni''<br />
| + | |
| - | **[[2xr8]] – BxBDO + Fe+2 + Fe2S2 <br />
| + | |
| - | **[[2xso]], [[2yfi]] – BxBDO (mutant) + Fe+2 + Fe2S2 <br />
| + | |
| - | **[[1uli]] - RoBDO + Fe+2 + Fe2S2 <br />
| + | |
| - | **[[2gbw]] - SyBDO + Fe+2 + Fe2S2 + O2 – ''Sphingobium yanoikuyae''<br />
| + | |
| - | | + | |
| - | *''BDO complex with aromatic compound''
| + | |
| - | | + | |
| - | **[[3gzx]] – CtBDO + Fe+2 + Fe2S2 + biphenyl <br />
| + | |
| - | **[[2xrx]] – BxBDO + Fe+2 + Fe2S2 + biphenyl <br />
| + | |
| - | **[[2xsh]] – BxBDO (mutant) + Fe+2 + Fe2S2 + biphenyl <br />
| + | |
| - | **[[2yfj]], [[2yfl]] – BxBDO (mutant) + Fe+2 + Fe2S2 + dibenzofuran <br />
| + | |
| - | **[[1ulj]] - RoBDO + Fe+2 + Fe2S2 + biphenyl <br />
| + | |
| - | **[[2gbx]] - SyBDO + Fe+2 + Fe2S2 + biphenyl <br />
| + | |
| - | | + | |
| - | *Tryptophan 2,3-dioxygenase (TDO)
| + | |
| - | | + | |
| - | **[[1yw0]] – XcTDO + Mg – ''Xanthomonas campestris''<br />
| + | |
| - | **[[2nw7]] – XcTDO + heme <br />
| + | |
| - | **[[4hka]] – TDO + heme – ''Drosophila melanogaster''<br />
| + | |
| - | **[[3bk9]], [[3e08]] – XcTDO (mutant) + heme + tryptophan <br />
| + | |
| - | **[[2nw8]], [[2nw9]] – XcTDO + heme + Mn + tryptophan derivative<br />
| + | |
| - | **[[2nox]] – CmTDO + heme – ''Cupriavidus metallidurans''<br />
| + | |
| - | **[[4pw8]] - hTDO + Co+2 <br />
| + | |
| - | | + | |
| - | *Cysteine dioxygenase type I (CysDO)
| + | |
| - | | + | |
| - | **[[2gh2]], [[4ieo]], [[4iep]], [[4ieq]], [[4ier]], [[4ies]], [[4iet]], [[4ieu]], [[4iev]], [[4iew]], [[4iex]], [[4iey]], [[4iez]], [[4jtn]], [[4jto]], [[4kwj]], [[4kwk]], [[4kwl]], [[2b5h]] – rCysDO + Fe+2 – rat <br />
| + | |
| - | **[[4ubg]], [[4ubh]] – rCysDO (mutant) + Fe+2 <br />
| + | |
| - | **[[3eln]] – rCysDO + Fe+2 + cysteine derivative<br />
| + | |
| - | **[[2q4s]], [[2atf]] – mCysDO + Ni+2 <br />
| + | |
| - | **[[2ic1]] – hCysDO + Fe+2 + cysteine<br />
| + | |
| - | | + | |
| - | *4-hydroxyphenylpyruvate dioxygenase (HPPD)
| + | |
| - | | + | |
| - | **[[1cjx]] – HPPD + Fe+2 – ''Pseudomonas fluorescens''<br />
| + | |
| - | **[[1t47]] – HPPD + Fe+2 – ''Streptomyces avermitilis''<br />
| + | |
| - | **[[3zgj]] – HPPD + Co+2 + mandelic acid – ''Streptomyces coelicolor''<br />
| + | |
| - | **[[1sqd]], [[1sp9]] – AtHPPD + Fe+3 <br />
| + | |
| - | **[[1sqi]] – HPPD + Fe+3 + inhibitor - rat<br />
| + | |
| - | **[[1tfz]], [[1tg5]] – AtHPPD + Fe+3 + inhibitor <br />
| + | |
| - | **[[3isq]] – hHPPD + Co+2 - human<br />
| + | |
| - | **[[1sp8]] – HPPD + Fe+2 – corn<br />
| + | |
| - | | + | |
| - | *Quercetin 2,3-dioxygenase (QDO)
| + | |
| - | | + | |
| - | **[[1juh]] – AjQDO + Cu+2 – ''Aspergillus japonicus''<br />
| + | |
| - | **[[2h0v]] – QDO + Fe+3 – ''Bacillus subtilis''<br />
| + | |
| - | **[[1gqg]], [[1gqh]] – AjQDO + Cu+2 + inhibitor<br />
| + | |
| - | **[[1h1i]] – AjQDO + Cu+2 + quercetin<br />
| + | |
| - | **[[1h1m]] – AjQDO + Cu+2 + kaempferol<br />
| + | |
| - | | + | |
| - | *Taurine/α-ketoglutarate dioxygenase (TauD)
| + | |
| - | | + | |
| - | **[[1gqw]], [[1gy9]], [[1os7]] – EcTauD + Fe+2 + taurine + oxoglutarate <br />
| + | |
| - | **[[1otj]] – EcTauD + taurine <br />
| + | |
| - | **[[3pvj]], [[3v15]] – PpTauD <br />
| + | |
| - | **[[3v17]] – PpTauD + oxoglutarate <br />
| + | |
| - | **[[3r1j]] – HAD + Fe+3 – ''Mycobacterium avium''<br />
| + | |
| - | **[[4j5i]] – HAD + Fe+3 – ''Mycobacterium smegmatis''<br />
| + | |
| - | **[[3swt]] – HAD + Fe+3 – ''Mycobacterium marinum''<br />
| + | |
| - | | + | |
| - | *3-hydroxyanthranilate 3,4-dioxygenase (HAD)
| + | |
| - | | + | |
| - | **[[1yfu]], [[4hvq]], [[4hvr]], [[4i3p]] – CmHAD + Fe+3<br />
| + | |
| - | **[[4hvo]] – CmHAD + Fe+3 + Cu+2<br />
| + | |
| - | **[[1yfw]] – CmHAD + Fe+3 + O2 + hydroxyanthranilate<br />
| + | |
| - | **[[1yfx]] – CmHAD + Fe+3 + NO + hydroxyanthranilate<br />
| + | |
| - | **[[1yfy]] – CmHAD + Fe+3 + hydroxyanthranilate<br />
| + | |
| - | **[[1zvf]] – HAD + Ni+2 - yeast<br />
| + | |
| - | **[[2qnk]] – hHAD + Ni+2 <br />
| + | |
| - | **[[3fe5]] – HAD + Fe+3 - bovine<br />
| + | |
| - | | + | |
| - | *Carbazole 1,9a-dioxygenase (CDO)
| + | |
| - | | + | |
| - | **[[1vck]] – CDO ferredoxin component + Fe+3 + Fe2S2 – ''Pseudomonas resinovorans''<br />
| + | |
| - | **[[2de5]], [[2de6]] – JaCDO ferredoxin and terminal oxygenase components + Fe+3 + Fe2S2 – ''Janthinobacterium''<br />
| + | |
| - | **[[2de7]], [[3vmg]] – JaCDO ferredoxin and terminal oxygenase components + Fe+3 + Fe2S2 + carbazole<br />
| + | |
| - | **[[3vmh]] – JaCDO ferredoxin and terminal oxygenase components + Fe+3 + Fe2S2 + O2<br />
| + | |
| - | **[[4nb8]], [[4nb9]], [[4nba]], [[4nbb]], [[4nbc]], [[4nbd]], [[4nbe]], [[4nbf]], [[4nbg]], [[4nbh]] – JaCDO ferredoxin and terminal oxygenase components (mutant) + Fe+2 + Fe2S2 <br />
| + | |
| - | **[[3vmi]] – JaCDO ferredoxin and terminal oxygenase components + Fe+3 + Fe2S2 + carbazole + O2<br />
| + | |
| - | **[[3gcf]] – CDO terminal oxygenase component + Fe+2 + Fe2S2 – ''Nocardioides aromaticivorans''<br />
| + | |
| - | **[[3gkq]] – SpCDO terminal oxygenase component + Fe+2 + Fe2S2 <br />
| + | |
| - | | + | |
| - | *Homogentisate 1,2-dioxygenase (HGDO)
| + | |
| - | | + | |
| - | **[[1ey2]] – hHGDO + Fe+2 <br />
| + | |
| - | **[[1eyb]] – hHGDO <br />
| + | |
| - | **[[4aq2]] – PpHGDO + Fe+3 <br />
| + | |
| - | **[[4aq6]] – PpHGDO + Fe+3 + substrate<br />
| + | |
| - | **[[3zds]] – PpHGDO + Fe+3 + O2<br />
| + | |
| - | | + | |
| - | *Hydroxyethylphosphonate dioxygenase (PHPD)
| + | |
| - | | + | |
| - | **[[4yar]], [[3g7d]] – SvPHPD + Cd+2 – ''Streptomyces viridochromogenes''<br />
| + | |
| - | **[[3rzz]] – SvPHPD (mutant) + Cd+2 <br />
| + | |
| - | **[[3gbf]] – SvPHPD + Cd+2 + hydroxyethylphosphonate <br />
| + | |
| - | | + | |
| - | *Gallate dioxygenase (GD)
| + | |
| - | | + | |
| - | **[[3wrc]] – SpGD + Fe+2 + dehydroxybenzoate<br />
| + | |
| - | **[[3wku]], [[3wpm]], [[3wr4]] – SpGD + Fe+3 + gallate<br />
| + | |
| - | **[[3wr3]], [[3wr9]] – SpGD + Fe+2 + gallate<br />
| + | |
| - | **[[3wrb]] – SpGD (mutant) + Fe+2 + gallate<br />
| + | |
| - | **[[3wr8]], [[3wra]] – SpGD + Fe+2<br />
| + | |
| - | | + | |
| - | * Glyoxalase/bleomycin resistant protein/dioxygenase (GBD)
| + | |
| - | | + | |
| - | **[[2rbb]] – GBD – ''Burkholderia phytofirmans''<br />
| + | |
| - | **[[2rk9]] – GBD – ''Vibrio splendidus''<br />
| + | |
| - | **[[3b59]] – GBD – ''Novosphingobium aromaticivorans''<br />
| + | |
| - | **[[3kol]] – GBD – ''Nostoc punctiforme''<br />
| + | |
| - | **[[3rri]] – GBD – ''Alicyclobacillus acidocaldarius''<br />
| + | |
| - | **[[3vb0]] – GBD – ''Sphingomonas wittichii''<br />
| + | |
| - | **[[3vcx]] – GBD – ''Pseudomonas palustris''<br />
| + | |
| - | **[[4g6x]] – CaGBD – ''Catenulispora acidiphila''<br />
| + | |
| - | **[[4n04]] – CaGBD (mutant)<br />
| + | |
| - | **[[4gym]] – GBD – ''Conexibacter woesei''<br />
| + | |
| - | **[[4hc5]] – GBD – ''Sphaerobacter thermophilus''<br />
| + | |
| - | **[[4mym]] – GBD – ''Nocardioides''<br />
| + | |
| - | **[[4qb5]] – GBD – ''Albidiferax ferrireducens''<br />
| + | |
| - | **[[4rt5]] – GBD – ''Planctomyces limnophilus''<br />
| + | |
| - | | + | |
| - | }}
| + | |
| | <references/> | | <references/> |
| | [[Category:Topic Page]] | | [[Category:Topic Page]] |