Acylaminoacyl peptidase
From Proteopedia
(Difference between revisions)
| (20 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='350' side='right' scene='43/433643/Cv/2' caption='Acylaminoacyl peptidase dimer complex with acetyl and glycerol [[2hu7]]'> | |
| - | + | ||
| + | == Function == | ||
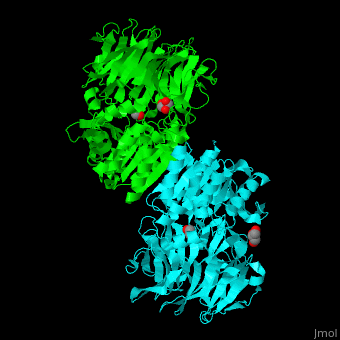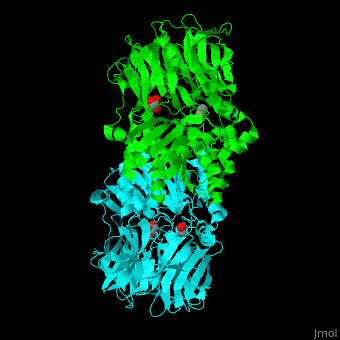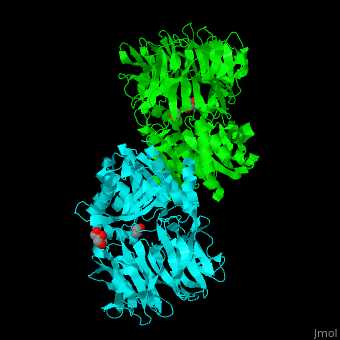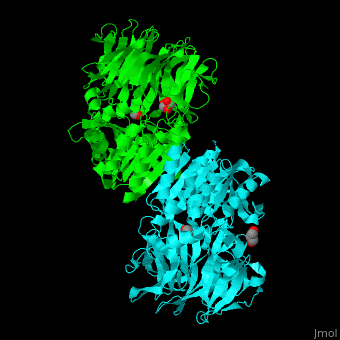
| + | [[Acylaminoacyl peptidase]] or '''Acylamino-acid releasing enzyme''' ('''AARE''', EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.4.19.1 3.4.19.1]) cleaves N-acetyl or N-formyl amino acid from the N-terminal of polypeptides.<ref>PMID:21084296</ref> | ||
| + | == Structural highlights == | ||
| + | AARE contains a <scene name='43/433643/Cv/6'>C-terminal hydrolase</scene> (in olive) and an <scene name='43/433643/Cv/7'>N-terminal β-propeller domain</scene> (in green). <scene name='43/433643/Cv/8'>Active site is S445, D524, H556</scene>. <ref>PMID:17350041</ref> The enzyme exists in an open state in which the oxyanion active site is accessible to the substrate and in a closed state where the active site is blocked. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | [[Acylaminoacyl peptidase]] or Acylamino-acid releasing enzyme ('''AARE''', EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.4.19.1 3.4.19.1]) cleaves N-acetyl or N-formyl amino acid from the N-terminal of polypeptides. The images at the left and at the right correspond to one representative AARE, ''i.e.'' the crystal structure of Acylaminoacyl peptidase from ''Aeropyrum pernix'' ([[1ve6]]). | ||
| - | |||
| - | {{TOC limit|limit=2}} | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
== 3D Structures of Acylaminoacyl peptidase == | == 3D Structures of Acylaminoacyl peptidase == | ||
| + | [[Acylaminoacyl peptidase 3D structures]] | ||
| + | </StructureSection> | ||
| + | __NOTOC__ | ||
| - | [[3o4g]], [[3o4h]], [[3o4i]], [[3o4j]], [[2hu5]], [[2hu7]], [[2hu8]], [[1ve6]] – ApAARE – ''Aeropyrum pernix''<br /> | ||
| - | [[2qr5]], [[2qzp]] – ApAARE (mutant)<br /> | ||
| - | [[1ve7]] – ApAARE+p-nitrophenyl phosphate<br /> | ||
| - | [[3fnb]] – AARE – ''Streptococcus mutans''<br /> | ||
| + | == References == | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Harmat V, Domokos K, Menyhard DK, Pallo A, Szeltner Z, Szamosi I, Beke-Somfai T, Naray-Szabo G, Polgar L. Structure and catalysis of acylaminoacyl peptidase: closed and open subunits of a dimer oligopeptidase. J Biol Chem. 2010 Nov 16. PMID:21084296 doi:10.1074/jbc.M110.169862
- ↑ Kiss AL, Hornung B, Radi K, Gengeliczki Z, Sztaray B, Juhasz T, Szeltner Z, Harmat V, Polgar L. The acylaminoacyl peptidase from Aeropyrum pernix K1 thought to be an exopeptidase displays endopeptidase activity. J Mol Biol. 2007 Apr 27;368(2):509-20. Epub 2007 Feb 20. PMID:17350041 doi:10.1016/j.jmb.2007.02.025