Apurinic-Apyrimidinic Endonuclease
From Proteopedia
(Difference between revisions)
m (Apurinic-Apyrimidinic Endonuclease-1 moved to Apurinic-Apyrimidinic Endonuclease: requested by Editor) |
|||
| (9 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' caption='Zebrafish APE1 trimer interacting with Pb+2 ions. [[2o3c]]' scene='37/377764/Cv/1'> |
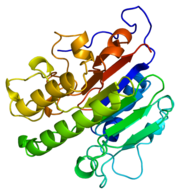
[[Image:thumbnail.png|thumb|left|Figure 1.Ribbon Structure of APE1. Antiparallel beta sheets line the interior of the enzyme, while alpha helices line the exterior.]] | [[Image:thumbnail.png|thumb|left|Figure 1.Ribbon Structure of APE1. Antiparallel beta sheets line the interior of the enzyme, while alpha helices line the exterior.]] | ||
==Function== | ==Function== | ||
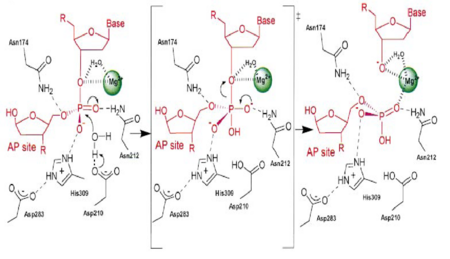
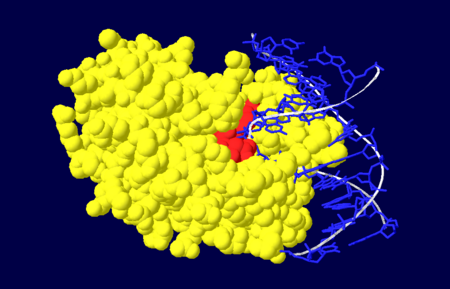
| - | DNA is damaged frequently in living cells via endogenous and exogenous factors that have the ability to cause abasic sites in the DNA strand. Abasic DNA must be repaired both accuratly and efficiently in order to prevent the death of the cell or the accumulation of mutations within the genome. Abasic DNA is repaired in the base excision repair pathway. '''Apurinic/apyrimidinic endonuclease-1''' (APE1) is the major repair enzyme in the base excision repair pathway in humans <ref name="Masood">Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879</ref>. APE1 has the ability to cleave the phophodiester bond at the 5' end of abasic sites in damaged DNA strands <ref name="Masood"/>. | + | DNA is damaged frequently in living cells via endogenous and exogenous factors that have the ability to cause abasic sites in the DNA strand. Abasic DNA must be repaired both accuratly and efficiently in order to prevent the death of the cell or the accumulation of mutations within the genome. Abasic DNA is repaired in the base excision repair pathway. '''Apurinic/apyrimidinic endonuclease-1''' (APE1) is the major repair enzyme in the base excision repair pathway in humans <ref name="Masood">Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879</ref>. APE1 has the ability to cleave the phophodiester bond at the 5' end of abasic sites in damaged DNA strands <ref name="Masood"/>. '''Apurinic/apyrimidinic endonuclease-2''' (APE2) protects B cells from oxidative damage <ref>PMID:24935922</ref>. '''Apurinic/apyrimidinic endonuclease-4''' (APE4) is present in lower organisms<ref>PMID:20667510</ref>. |
An important point of gene regulation within a cell is the regulation of the half life of mRNA transcripts. The regulation of mRNA transcripts will control the amount of protein translated in a cell. Many mRNA transcripts have been linked to diseases such as [[cancer]]. These deleterious mRNA transcripts are known as proto-oncogene products and can greatly effect the health of cells<ref name="Barnes"> Barnes, T., et al. 2009. Identification of Apurinic/apyrimidinic endoribonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA in vitro. Nucleic Acid Research 37: 3946-3958</ref>. APE1 has also been shown to have the ability to regulate the half life of c-''myc'' mRNA by endonucleolytically cleaving the coding region determinant (CRD) of c-''myc'' mRNA<ref name="Barnes"/>. | An important point of gene regulation within a cell is the regulation of the half life of mRNA transcripts. The regulation of mRNA transcripts will control the amount of protein translated in a cell. Many mRNA transcripts have been linked to diseases such as [[cancer]]. These deleterious mRNA transcripts are known as proto-oncogene products and can greatly effect the health of cells<ref name="Barnes"> Barnes, T., et al. 2009. Identification of Apurinic/apyrimidinic endoribonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA in vitro. Nucleic Acid Research 37: 3946-3958</ref>. APE1 has also been shown to have the ability to regulate the half life of c-''myc'' mRNA by endonucleolytically cleaving the coding region determinant (CRD) of c-''myc'' mRNA<ref name="Barnes"/>. | ||
| Line 34: | Line 34: | ||
* [http://www.rcsb.org/pdb/explore/explore.do?structureId=1DE8 APE1 (1de8) RCSB PDB page] | * [http://www.rcsb.org/pdb/explore/explore.do?structureId=1DE8 APE1 (1de8) RCSB PDB page] | ||
* [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=4.2.99.18 BRENDA-The comprehensive enzyme information system-APE1 page] | * [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=4.2.99.18 BRENDA-The comprehensive enzyme information system-APE1 page] | ||
| - | </StructureSection> | ||
==3D structures of Apurinic/Apyrimidinic Endonuclease== | ==3D structures of Apurinic/Apyrimidinic Endonuclease== | ||
| + | [[Apurinic/apyrimidinic endonuclease 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==References== | ==References== | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879
- ↑ Guikema JE, Linehan EK, Esa N, Tsuchimoto D, Nakabeppu Y, Woodland RT, Schrader CE. Apurinic/apyrimidinic endonuclease 2 regulates the expansion of germinal centers by protecting against activation-induced cytidine deaminase-independent DNA damage in B cells. J Immunol. 2014 Jul 15;193(2):931-9. doi: 10.4049/jimmunol.1400002. Epub 2014 Jun, 16. PMID:24935922 doi:http://dx.doi.org/10.4049/jimmunol.1400002
- ↑ Daley JM, Zakaria C, Ramotar D. The endonuclease IV family of apurinic/apyrimidinic endonucleases. Mutat Res. 2010 Dec;705(3):217-27. doi: 10.1016/j.mrrev.2010.07.003. Epub 2010, Aug 3. PMID:20667510 doi:http://dx.doi.org/10.1016/j.mrrev.2010.07.003
- ↑ 4.0 4.1 Barnes, T., et al. 2009. Identification of Apurinic/apyrimidinic endoribonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA in vitro. Nucleic Acid Research 37: 3946-3958
- ↑ 5.0 5.1 5.2 Ando, K. et al. 2008. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kB and AP-1, and activation of their DNA binding activity. Nucleic Acid research 36(13): 4327-4336.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 6.8 6.9 Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.
Proteopedia Page Contributors and Editors (what is this?)
Mark Barnes, Michal Harel, Alexander Berchansky, David Canner, Jaime Prilusky, Andrea Gorrell, Eran Hodis, Joel L. Sussman