We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Cystine
From Proteopedia
(Difference between revisions)
m (New page: Cystine is formed by the oxidation of two proximal cysteine residues, covalently linking them in a disulfide bond. [[Image:1ogscystines.png|left|thumb|360px|<span style="font-size:1.2...) |
('&' in Structure tag caption was causing XML error. Replaced it with 'and'.) |
||
| (13 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
[[Image:1ogscystines.png|left|thumb|360px|<span style="font-size:1.2em;">Two cystines in human acid-beta-glucosidase (GlcCerase), the enzyme mutated in [[Treatment of Gaucher disease|Gaucher disease]], from [[1ogs]].</span>]] | [[Image:1ogscystines.png|left|thumb|360px|<span style="font-size:1.2em;">Two cystines in human acid-beta-glucosidase (GlcCerase), the enzyme mutated in [[Treatment of Gaucher disease|Gaucher disease]], from [[1ogs]].</span>]] | ||
| - | < | + | <Structure load='1ogs' frame='true' align='right' size='350' |
| + | caption='Disulfide Connectivity of Human beta-glucocerebrosidase, Cys4-Cys16 and Cys18-Cys23 ([[1ogs]])' scene='User:Adriana_Kita/Sandbox_1/Gcase_disulfide_1/2'/> | ||
| - | + | ||
| - | + | ==Visualization== | |
| + | [[FirstGlance in Jmol]] displays all cystines (disulfide bonds) in any given protein in a clear manner. To display a protein in ''FirstGlance'' and see its disulfide bonds: | ||
| + | *Find a page in Proteopedia about that protein. You can use the search slot at left to search for the name of the protein, or a specific [[PDB code]]. | ||
| + | *If the page is not titled with a PDB code, look for mention of a PDB code, and go to the page titled with that PDB code. | ||
| + | *Click on the link ''FirstGlance'' beneath the molecule. | ||
| + | *Once you see the protein in ''FirstGlance'', click on ''More Views..'', then on ''Disulfide bonds: show all''. | ||
| + | |||
| + | Eventually, you will be able to select and display disulfide bonds in Proteopedia's [[Scene Authoring Tools]] (SAT). In March, 2012, this is on the [[Wishlist#Wishlist_for_Inside_the_Molecular_Scene_Authoring_Tools_.28SAT.29|Wishlist]]. Currently, in the [[SAT]], you can select CYS, which includes both cysteine and cystine. | ||
| + | |||
| + | ==Articles== | ||
Articles in Proteopedia concerning cystine include: | Articles in Proteopedia concerning cystine include: | ||
*[[Disulfide Connectivity of Velaglucerase|Disulfide Connectivity of Velaglucerase, a.k.a. acid-beta-glucosidase (GlcCerase)]] | *[[Disulfide Connectivity of Velaglucerase|Disulfide Connectivity of Velaglucerase, a.k.a. acid-beta-glucosidase (GlcCerase)]] | ||
*[[Serine Proteases]] | *[[Serine Proteases]] | ||
*[[PHB synthase in Rhodobacter sphaeroides]] | *[[PHB synthase in Rhodobacter sphaeroides]] | ||
| - | *[[Serine Proteases: A Tutorial of Chymotrypsin, Trypsin and Elastase]] | ||
*[[User:Wayne Decatur/Alter Disulfide Bonds in a Structure|Altering Disulfide Bonds in a Structure Using PyMol]] | *[[User:Wayne Decatur/Alter Disulfide Bonds in a Structure|Altering Disulfide Bonds in a Structure Using PyMol]] | ||
| - | <br /> | ||
| - | To view automatically seeded indices concerning cystine, see: | + | ==Cystine Categories== |
| + | To view automatically seeded "Category" indices concerning cystine, see: | ||
*[[:Category:Cyclic cystine knot|Cyclic cystine knot]] | *[[:Category:Cyclic cystine knot|Cyclic cystine knot]] | ||
*[[:Category:Cystine knot superfamily|Cystine knot superfamily]] | *[[:Category:Cystine knot superfamily|Cystine knot superfamily]] | ||
| Line 34: | Line 43: | ||
==References and Notes== | ==References and Notes== | ||
| - | <references/> | + | <references /> |
==See Also== | ==See Also== | ||
*[[Disulfide bond]] | *[[Disulfide bond]] | ||
*[[Metabolic Disorders ]] | *[[Metabolic Disorders ]] | ||
| + | *[[Selenocysteine]] | ||
==Additional Literature== | ==Additional Literature== | ||
| Line 46: | Line 56: | ||
*[http://clavius.bc.edu/~clotelab/DiANNA/ DiANNA: a web server for disulfide connectivity prediction] | *[http://clavius.bc.edu/~clotelab/DiANNA/ DiANNA: a web server for disulfide connectivity prediction] | ||
*[http://disulfind.dsi.unifi.it/ DISULFIND: Cysteines Disulfide Bonding State and Connectivity Predictor] | *[http://disulfind.dsi.unifi.it/ DISULFIND: Cysteines Disulfide Bonding State and Connectivity Predictor] | ||
| + | [[Category:Topic Page]] | ||
Current revision
Cystine is formed by the oxidation of two proximal cysteine residues, covalently linking them in a disulfide bond.
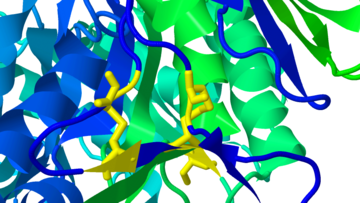
Two cystines in human acid-beta-glucosidase (GlcCerase), the enzyme mutated in Gaucher disease, from 1ogs.
|
Contents |
Visualization
FirstGlance in Jmol displays all cystines (disulfide bonds) in any given protein in a clear manner. To display a protein in FirstGlance and see its disulfide bonds:
- Find a page in Proteopedia about that protein. You can use the search slot at left to search for the name of the protein, or a specific PDB code.
- If the page is not titled with a PDB code, look for mention of a PDB code, and go to the page titled with that PDB code.
- Click on the link FirstGlance beneath the molecule.
- Once you see the protein in FirstGlance, click on More Views.., then on Disulfide bonds: show all.
Eventually, you will be able to select and display disulfide bonds in Proteopedia's Scene Authoring Tools (SAT). In March, 2012, this is on the Wishlist. Currently, in the SAT, you can select CYS, which includes both cysteine and cystine.
Articles
Articles in Proteopedia concerning cystine include:
- Disulfide Connectivity of Velaglucerase, a.k.a. acid-beta-glucosidase (GlcCerase)
- Serine Proteases
- PHB synthase in Rhodobacter sphaeroides
- Altering Disulfide Bonds in a Structure Using PyMol
Cystine Categories
To view automatically seeded "Category" indices concerning cystine, see:
- Cyclic cystine knot
- Cystine knot superfamily
- Cystine knot
- Cystine stabilized alpha-beta motif
- Inhibitor cystine knot
- Cystine-stabilized alpha-helical motif
- Cystine-knot
- Cystine-rich
- Cystine-knot growth factor
- Inhibitor cystine knot motif
- Inhibitor cystine-knot
- Cyclic cystine knot motif
- Homodimer,cystine knot
- Inhibitory cystine knot
- Fad-cystine-oxidoreductase
- Cystine knot motif
References and Notes
See Also
Additional Literature
- Ladenstein R, Ren B. Reconsideration of an early dogma, saying "there is no evidence for disulfide bonds in proteins from archaea". Extremophiles. 2008 Jan;12(1):29-38. Epub 2007 May 17. PMID:17508126 doi:10.1007/s00792-007-0076-z
- Dvir H, Harel M, McCarthy AA, Toker L, Silman I, Futerman AH, Sussman JL. X-ray structure of human acid-beta-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003 Jul;4(7):704-9. PMID:12792654 doi:10.1038/sj.embor.embor873
